Hi everyone,
I am currently working on the characterization of my chromatographic system fitting CSTR and DPFR parameters to describe BTCs under non-binding conditions (system only, no column attached yet), very similar to this script:
https://cadet-process.readthedocs.io/en/latest/examples/characterize_chromatographic_system/column_transport_parameters.html
Everthing seems fine until the optimization using U_NSGA3 starts. Here, I receive an error message:
AttributeError: ‘Cadet’ object has no attribute ‘_is_file_class’
Something seems not to be defined properly in CADET. Does someone have experience with this? Thanks in advance!
Best,
Nils
Hi Nils,
could you please also let us know on which OS you’re working, give us a dump of the package versions in your python environment and if you’re using Jupyter Notebooks / Lab to execute the code?
Thanks!
Looked into it a bit more today and I can’t solve it right now. It appears to be a pickling issue with ipython.
The workaround that works for me is to run the script with regular python instead of ipython. So in a console run $python “path/to/your/script.py”.
Hi Nils,
We have been working on a fix that hasn’t made it into the stable release yet. You can try the version of CADET-Process which includes the fix with this pip command:
pip install -e git+https://github.com/fau-advanced-separations/CADET-Process.git@add_selector_for_parallelization_backend#egg=CADET-Process
Hi Ron,
sorry for the late reply, vacation season 
Is this website still available?
pip install -e git+https://github.com/fau-advanced-separations/CADET-Process.git@add_selector_for_parallelization_backend#egg=CADET-Process
I can’t find anything there.
Best
Nils
Hey Nils,
nice, hope you had a nice vacation!
The parallelization branch has been merged into the dev branch, but not into the master branch yet, so the current pip command is
pip install -e git+https://github.com/fau-advanced-separations/CADET-Process.git@dev#egg=CADET-Process
Just ran it in a fresh conda env to test it and it worked for me.
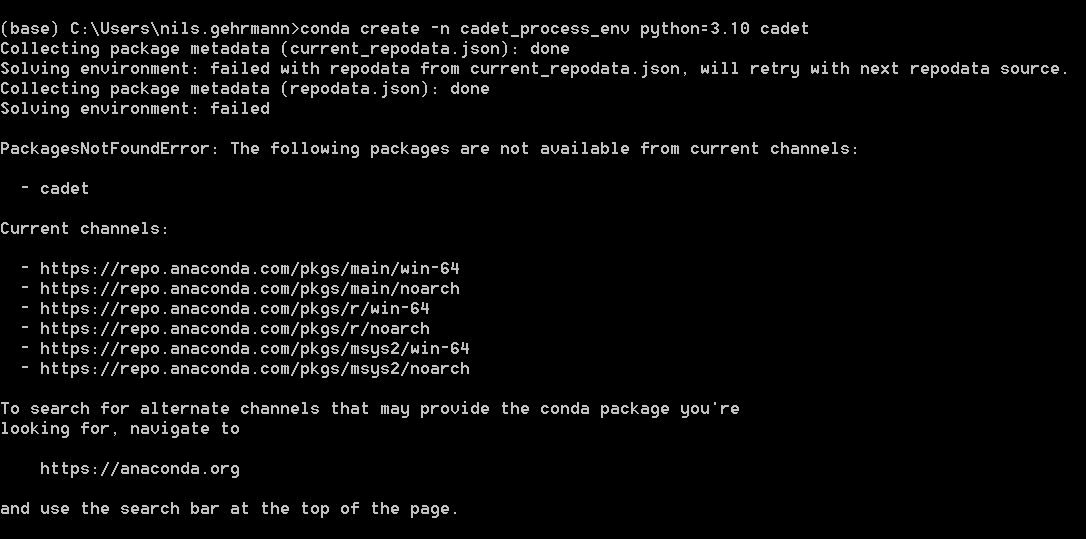
conda create -n cadet_process_env python=3.10 cadet
conda activate cadet_process_env
pip install -e git+https://github.com/fau-advanced-separations/CADET-Process.git@dev#egg=CADET-Process
Hi Ron,
yes, it was really good and refreshing, thank you 
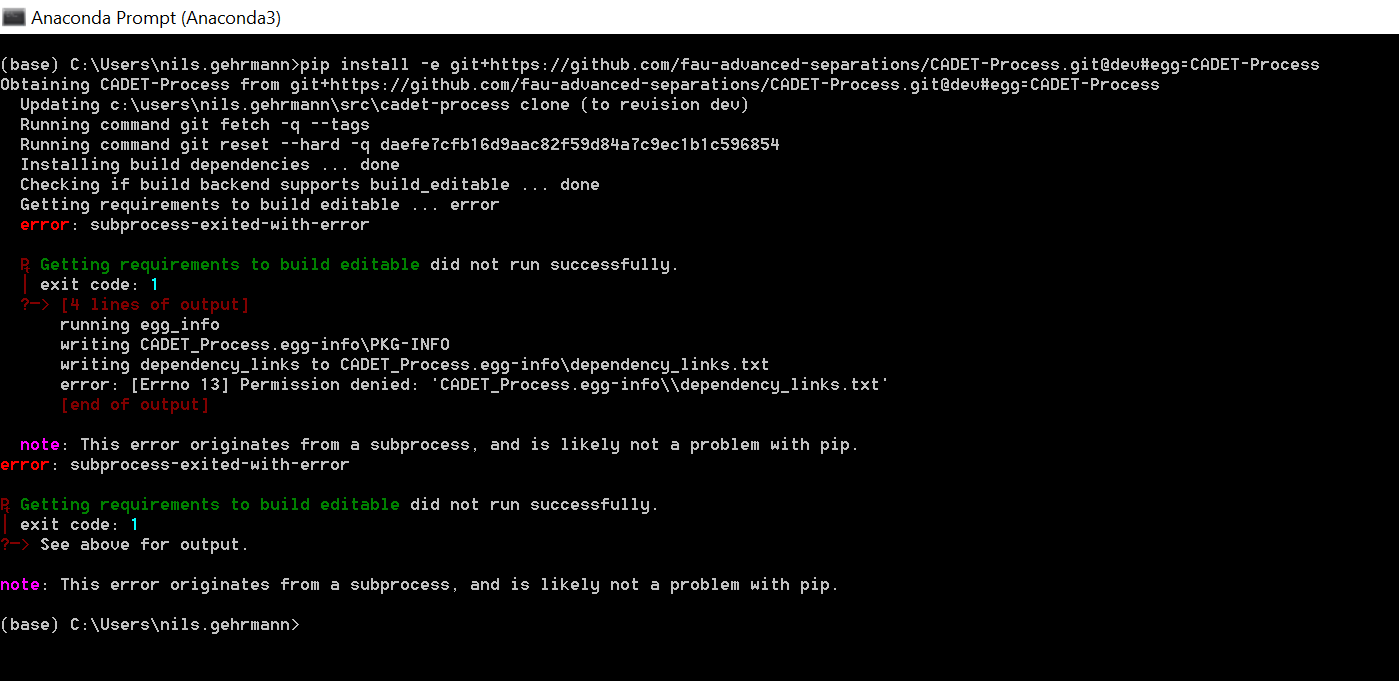
Unfortunately, the modified installation does not work for me :
Followingly, I am also not able to create the new environment:
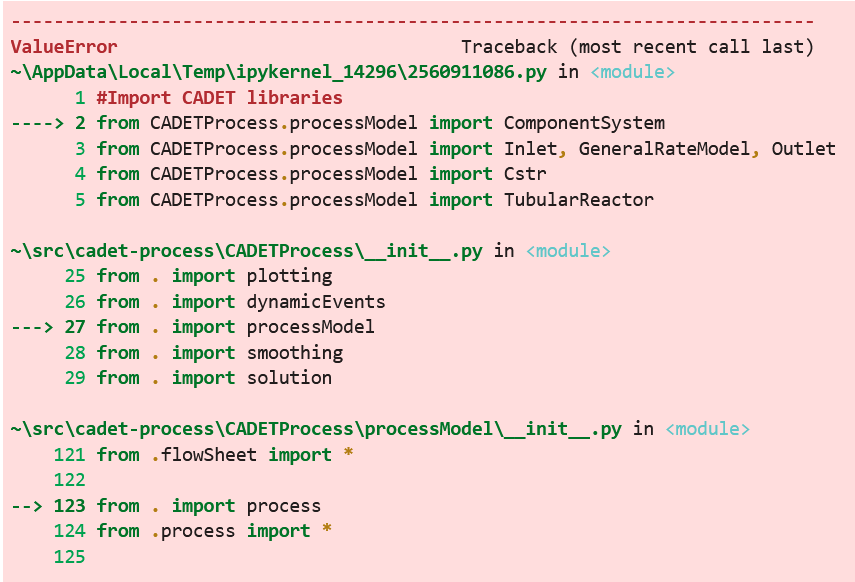
This results in my scripts not being able to import the process model:
What am I doing wrong?
Best
Nils
Hey Nils,
I’m really sorry, but I think that crashed your conda env. That happened to me too once. It’s a consequence of mixing conda with pip, which is fine most of the time, but can cause issues.
Because that command was executed in the base conda env I don’t know if that is recoverable or if you need to reinstall conda. Sorry about that!
The pip install -e etc. command should i.m.o only be run in a conda env, not in the base env. Sorry that I didn’t make that more clear in my message.
This Wednesday at 15:00 is the cadet office hour, if you want to go through the installation process together you can join us there.
So, we have a temporary fix: downgrade addict to v2.3
You can install cadet-process with addict 2.3 with this command from your cadet_process_env in conda:
pip install -e git+https://github.com/fau-advanced-separations/CADET-Process.git@fix/change_addict_version#egg=CADET-Process
Let me know if that works.
Hi Ron,
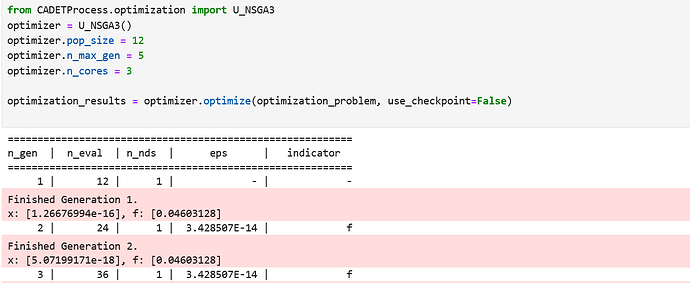
I just ran it with a small pop_size and yes, it works!
Thank you so much for your help!
2 Likes