Hello!
Can someone explain how to get mole balance between mobile phase and solid adsorbed.
I run ‘GENERAL_RATE_MODEL’ with ‘MULTI_COMPONENT_LANGMUIR’ adsorption model with zero flow rate as I need to fit our experimental data and find par_diffusion coeffitiant - it all works well except that amount of mole of adsorbed component on solid part after 300 minutes and what is left in liquid part is not equal to amount I started with at time zero.
I pull out solution_bulk, solution_solid and solution_particle, please see below
model.root.input[‘return’].unit_000.write_solution_solid = 1
model.root.input[‘return’].unit_000.write_solution_particle =1
model.root.input[‘return’].unit_000.write_solution_bulk = 1
and I expect that
initial solution_bulk * solution volume = solution_bulk * solution volume + solution_particle * pore volume + solution_solid * solid volume,
but it is not.
Please let me know where I am wrong.
Please see my script below:
import numpy as np
import matplotlib.pyplot as plt
from cadet import Cadet
Cadet.cadet_path = ‘C:\Users\konstantin\Anaconda3\pkgs\cadet-4.3.0-hbe28382_2\bin’
total_time =300*60 #s
time_frame = 1000
does not metter as flow rate is zero
col_length = 0.01 # m
cross_section_area = 0.1 # m2
col_porosity = 0.8
par_porosity = 0.1
par_radius = 3.25E-04 #m
col_dispersion = 3.789E-06 #m2/s
film_diffusion = [6.034E-02] #m/s
par_diffusion = [1.62E-09*8,] # m2/s
mcl_ka = [10.58735191,] # it is Langmuir coefficient b, m3/mol
mcl_kd = [1,]
mcl_qmax = [22471.33561/mcl_ka[0],] #mol / m^3
#Case A
concentration_ini = 1 #initial concentration in the solution mol/m3
init_q = 0
const_coeff = [concentration_ini,] # concentration mol/m3
Q = 0 # Flow rate in m^3*s^-1
#Model
model = Cadet()
model.root.input.model.nunits = 3
#Inlet Model
model.root.input.model.unit_000.unit_type = ‘INLET’
model.root.input.model.unit_000.ncomp = 1
model.root.input.model.unit_000.inlet_type = ‘PIECEWISE_CUBIC_POLY’
#General Rate Model
model.root.input.model.unit_001.unit_type = ‘GENERAL_RATE_MODEL’
model.root.input.model.unit_001.ncomp = 1
Geometry
model.root.input.model.unit_001.col_length = col_length
model.root.input.model.unit_001.cross_section_area = cross_section_area # m^2,
#model.root.input.model.unit_001.velocity = 0.000204933 # m/s
model.root.input.model.unit_001.col_porosity = col_porosity # -
model.root.input.model.unit_001.par_porosity = par_porosity # -
model.root.input.model.unit_001.par_radius = par_radius # m
Transport
model.root.input.model.unit_001.col_dispersion = col_dispersion # m^2 / s (interstitial volume)
model.root.input.model.unit_001.film_diffusion = film_diffusion # m / s
model.root.input.model.unit_001.par_diffusion = par_diffusion # m^2 / s (mobile phase)
model.root.input.model.unit_001.par_surfdiffusion = [0.0,] # zero m^2 / s (solid phase)
Adsorption Model
model.root.input.model.unit_001.adsorption_model = ‘MULTI_COMPONENT_LANGMUIR’
model.root.input.model.unit_001.adsorption.is_kinetic = True
model.root.input.model.unit_001.adsorption.mcl_ka = mcl_ka # m^3 / (mol * s) (mobile phase)
model.root.input.model.unit_001.adsorption.mcl_kd = mcl_kd # 1 / s (desorption)
model.root.input.model.unit_001.adsorption.mcl_qmax = mcl_qmax # mol / m^3 (solid phase)
Initial Conditions
model.root.input.model.unit_001.init_c = [concentration_ini,]
model.root.input.model.unit_001.init_q = [init_q,]
#Setting up the Discretization
Grid cells
model.root.input.model.unit_001.discretization.ncol = 100
model.root.input.model.unit_001.discretization.npar = 6
Bound states
model.root.input.model.unit_001.discretization.nbound = [1]
Other options
model.root.input.model.unit_001.discretization.par_disc_type = ‘EQUIDISTANT_PAR’
model.root.input.model.unit_001.discretization.use_analytic_jacobian = 1
model.root.input.model.unit_001.discretization.reconstruction = ‘WENO’
model.root.input.model.unit_001.discretization.gs_type = 1
model.root.input.model.unit_001.discretization.max_krylov = 0
model.root.input.model.unit_001.discretization.max_restarts = 10
model.root.input.model.unit_001.discretization.schur_safety = 1.0e-8
model.root.input.model.unit_001.discretization.weno.boundary_model = 0
model.root.input.model.unit_001.discretization.weno.weno_eps = 1e-10
model.root.input.model.unit_001.discretization.weno.weno_order = 3
Outlet Model
model.root.input.model.unit_002.unit_type = ‘OUTLET’
model.root.input.model.unit_002.ncomp = 1
#Time Sections
model.root.input.solver.sections.nsec = 1
model.root.input.solver.sections.section_times = [0.0,total_time,] # s
model.root.input.solver.sections.section_continuity = []
model.root.input.model.unit_000.sec_000.const_coeff = const_coeff # mol / m^3
model.root.input.model.unit_000.sec_000.lin_coeff = [0.0,]
model.root.input.model.unit_000.sec_000.quad_coeff = [0.0,]
model.root.input.model.unit_000.sec_000.cube_coeff = [0.0,]
#System Connectivity
model.root.input.model.connections.nswitches = 1
model.root.input.model.connections.switch_000.section = 0
model.root.input.model.connections.switch_000.connections = [
0, 1, -1, -1, Q, # [unit_000, unit_001, all components, all components, Q/ m^3s^-1
1, 2, -1, -1, Q] # [unit_001, unit_002, all components, all components, Q/ m^3s^-1
#Setting Up the Simulator and Running the Simulation
model.root.input.model.solver.gs_type = 1
model.root.input.model.solver.max_krylov = 0
model.root.input.model.solver.max_restarts = 10
model.root.input.model.solver.schur_safety = 1e-8
Number of cores for parallel simulation
model.root.input.solver.nthreads = 1
Tolerances for the time integrator
model.root.input.solver.time_integrator.abstol = 1e-6
model.root.input.solver.time_integrator.algtol = 1e-10
model.root.input.solver.time_integrator.reltol = 1e-6
model.root.input.solver.time_integrator.init_step_size = 1e-6
model.root.input.solver.time_integrator.max_steps = 1000000
#Return data
model.root.input[‘return’].split_components_data = 0
model.root.input[‘return’].split_ports_data = 0
model.root.input[‘return’].unit_000.write_solution_bulk = 1
model.root.input[‘return’].unit_000.write_solution_inlet = 1
model.root.input[‘return’].unit_000.write_solution_outlet = 1
model.root.input[‘return’].unit_000.write_coordinates = 1
model.root.input[‘return’].unit_000.write_solution_solid = 1 # return concentration on solid mol/m3
model.root.input[‘return’].unit_000.write_solution_particle =1
model.root.input[‘return’].unit_000.write_solution_volume = 1
Copy settings to the other unit operations
model.root.input[‘return’].unit_001 = model.root.input[‘return’].unit_000
model.root.input[‘return’].unit_002 = model.root.input[‘return’].unit_000
Solution times
model.root.input.solver.user_solution_times = np.linspace(0, total_time, 1001)
exec(open(‘utils.py’).read())
run_simulation(model)
Plotting the Results
plt.figure()
time = model.root.output.solution.solution_times
c = model.root.output.solution.unit_001.solution_inlet
c_max = np.max(c)
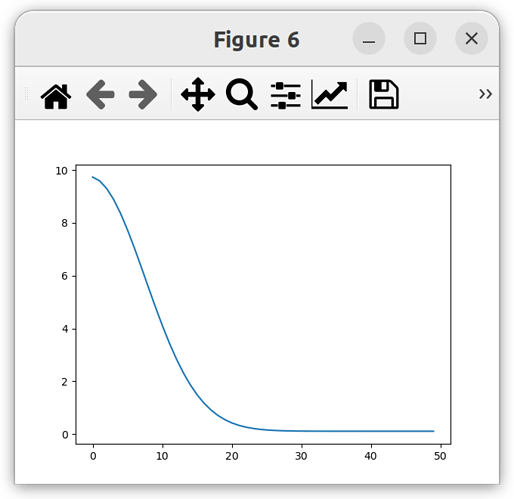
solution_solid = model.root.output.solution.unit_001.solution_solid[:, :, 0, 0]
solution_particle = model.root.output.solution.unit_001.solution_particle[:, :, 0, 0]
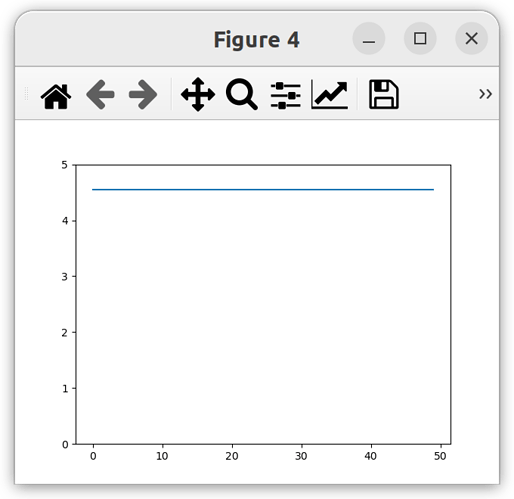
solution_volume = model.root.output.solution.unit_001.solution_volume
Thanks a lot!
Konstantin