Hi There,
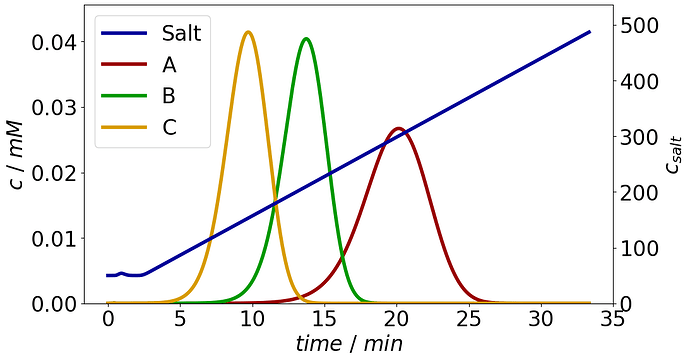
In the provided CADET Process example for bind and elute using the SMA model (figure below), what is the best way to combine the profiles for species A,B and C into a single “total concentration” output that is the additive combination of all 3 components.
For experimental data of more complex feedstocks (e.g. host cell proteins ) where I have multiple species but cannot measure the individual concentration only the additive effect of them all (e.g. a UV profile), I want to generate a simulation result that gives the “total concentration” profile of all my species combined which I can then be used as the object for optimisation against the raw experimental data, assuming the number of components is known.
Thanks
Will
Hey @Will_m32 and welcome to the forum!
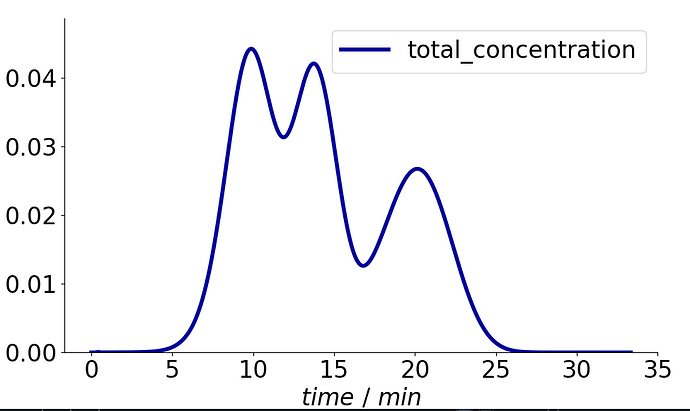
The simplest thing you could to is to use the slice_solution method.
It expects a solution object and allows you to specify flags such as use_total_concentration (which sums up all components) and components (a list of components you want to sum up).
In your example, this could look something like this:
from CADETProcess.solution import slice_solution
solution_new = slice_solution(simulation_results.solution.column.outlet, use_total_concentration=True, components=['A', 'B', 'C'])
solution_new.plot()
Which produces the following figure:
Note that I excluded the Salt component for the new solution object.
Also note that both the Comparator, as well as the Fractionator already support slicing so you can add the components list and use_total_concentration arguments in the corresponding methods.
1 Like
Hi there,
I had an additional question along these lines. If I have some data that is “total_concentration” data that I want to compare against the total_concentraiton simulation , how do i set up my component system for the Reference I0.
In my comparator I have the following system I want to compare. That is some protein data that is the sum of components A and B against the results from the simulation for total_concentration of ‘A’ and ‘B’.
comparator = Comparator()
comparator.add_reference(total_protein_data)
comparator.add_difference_metric(
'NRMSE', total_protein_data, 'outlet.outlet', use_total_concentration=True, components=['A','B'])
But I’m struggling to set up my reference I0 correctly. I need to assign components A and B to the data so that it is recognised by the comparator. However if I add components A and B to the Reference I0 it seems to expect two columns , one for each component.
file_path = 'path_to_excel.xlsx'
df_1 = pd.read_excel(file_path, skiprows=1, usecols=[0, 1])
df_1.columns = ['Time', 'protein']
protein_signal=df_1['protein'].to_numpy()
time =df_1['Time'].to_numpy()
component_system = ComponentSystem()
component_system.add_component('A')
component_system.add_component('B')
total_protein_data = ReferenceIO('protein',time ,protein_signal,component_system=component_system )
What’s the best way to approach this ?
Thanks
Hey,
so, the comparator expects to be able to do the same “processing” to both the reference and the simulation results. So your reference needs to have two components (A & B), so that the slicer can slice and add them together.
So if you have your experimental data in two numpy arrays you can construct a reference that contains both like this:
data_protein_A = np.array(data for protein A)
data_protein_B = np.array(data for protein B)
data_protein_stacked = np.stack([data_protein_A, data_protein_B], axis=-1)
reference1 = ReferenceIO(
f'Peak {file_name} - Component {component_index2}',
time_experiment, data_protein_stacked,
component_system=ComponentSystem(["A", "B"])
)