Hello, I am trying to set up an inlet profile in CADET-Process but running into an error.
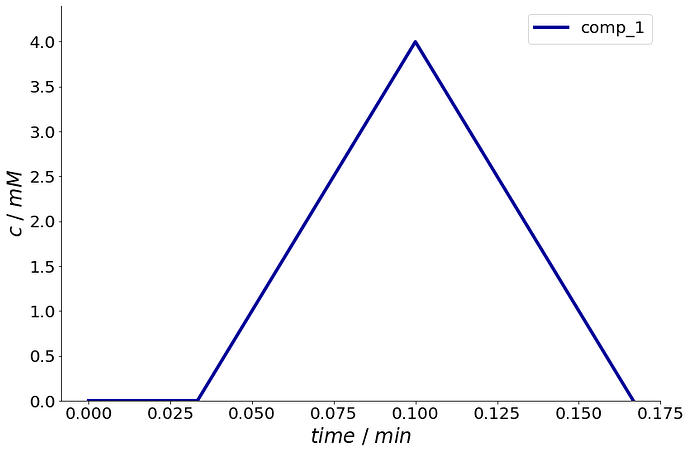
As a test case, what I am trying to do is set up the equivalent to the below, which gives the expected outlet plot:
from CADETProcess.processModel import ComponentSystem
from CADETProcess.processModel import Inlet, Outlet
from CADETProcess.processModel import FlowSheet, Process
from CADETProcess.simulator import Cadet
component_system = ComponentSystem(['comp_1'])
feed = Inlet(component_system, name='feed')
feed.flow_rate = 1e-6
outlet = Outlet(component_system, name='outlet')
flow_sheet = FlowSheet(component_system)
flow_sheet.add_unit(feed)
flow_sheet.add_unit(outlet)
flow_sheet.add_connection(feed, outlet)
process = Process(flow_sheet, 'test_process')
process.cycle_time = 10
_ = process.add_event('1', 'flow_sheet.feed.c', [0], 0)
_ = process.add_event('2', 'flow_sheet.feed.c', [0], 1)
_ = process.add_event('3', 'flow_sheet.feed.c', [1], 2)
_ = process.add_event('4', 'flow_sheet.feed.c', [2], 3)
_ = process.add_event('5', 'flow_sheet.feed.c', [3], 4)
_ = process.add_event('6', 'flow_sheet.feed.c', [4], 5)
_ = process.add_event('7', 'flow_sheet.feed.c', [3], 6)
_ = process.add_event('8', 'flow_sheet.feed.c', [2], 7)
_ = process.add_event('9', 'flow_sheet.feed.c', [1], 8)
_ = process.add_event('10', 'flow_sheet.feed.c', [0], 9)
# RUN SIMULATION
simulator = Cadet()
simulation_results = simulator.simulate(process)
_ = simulation_results.solution.outlet.inlet.plot()
If I am understanding correctly, I should be able to set up the same using the add_concentration_profile method as:
import numpy as np
from CADETProcess.processModel import ComponentSystem
from CADETProcess.processModel import Inlet, Outlet
from CADETProcess.processModel import FlowSheet, Process
from CADETProcess.simulator import Cadet
component_system = ComponentSystem(['comp_1'])
feed = Inlet(component_system, name='feed')
feed.flow_rate = 1e-6
outlet = Outlet(component_system, name='outlet')
flow_sheet = FlowSheet(component_system)
flow_sheet.add_unit(feed)
flow_sheet.add_unit(outlet)
flow_sheet.add_connection(feed, outlet)
process = Process(flow_sheet, 'test_process')
process.cycle_time = 10
# setting up inlet profile
time = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9,])
c = np.array([0, 0, 1, 2, 3, 4, 3, 2, 1, 0,])
process.add_concentration_profile(unit='feed', time=time, c=c)
# RUN SIMULATION
simulator = Cadet()
simulation_results = simulator.simulate(process)
_ = simulation_results.solution.outlet.inlet.plot()
But running this I get the error
File ~/Documents/000_code/CADET-Process_proA_model/CADET_Process_test.py:31
process.add_concentration_profile(unit='feed', time=time, c=c)
File ~/miniconda3/envs/CADET-env/lib/python3.12/site-packages/CADETProcess/processModel/process.py:578 in add_concentration_profile
elif components is None and c.shape[1] != self.n_comp:
IndexError: tuple index out of range
If I try to specify the component as
process.add_concentration_profile(unit='feed', time=time, c=c, components=[0,])
I get a different error:
File ~/Documents/000_code/CADET-Process_proA_model/CADET_Process_test.py:31
process.add_concentration_profile(unit='feed', time=time, c=c, components=[0,])
File ~/miniconda3/envs/CADET-env/lib/python3.12/site-packages/CADETProcess/processModel/process.py:598 in add_concentration_profile
evt = self.add_event(
File ~/miniconda3/envs/CADET-env/lib/python3.12/site-packages/CADETProcess/dynamicEvents/event.py:168 in add_event
evt = Event(name, self, parameter_path, state, time=time, indices=indices)
File ~/miniconda3/envs/CADET-env/lib/python3.12/site-packages/CADETProcess/dynamicEvents/event.py:889 in __init__
self.indices = indices
File ~/miniconda3/envs/CADET-env/lib/python3.12/site-packages/CADETProcess/dynamicEvents/event.py:1031 in indices
raise e
File ~/miniconda3/envs/CADET-env/lib/python3.12/site-packages/CADETProcess/dynamicEvents/event.py:1029 in indices
_ = self.indices
File ~/miniconda3/envs/CADET-env/lib/python3.12/site-packages/CADETProcess/dynamicEvents/event.py:1003 in indices
indices = generate_indices(self.parameter_shape, self._indices)
File ~/miniconda3/envs/CADET-env/lib/python3.12/site-packages/CADETProcess/dynamicEvents/section.py:682 in generate_indices
indices_array = np.array(indices, ndmin=1)
ValueError: invalid __array_struct__
Not sure if I am setting something up incorrectly or if there is a bug.
Thanks in advance for your help ![]()