Hello everyone,
I am experiencing a weird issue and cannot quite explain what is causing it. As alsready stated in another thread I am using Cubic splines to get Gaussian inlet profiles.
It is defined as
def get_spline_as_input(cadet, gauss_t, t, y):
cadet.root.input.model.unit_000.unit_type = 'INLET'
cadet.root.input.model.unit_000.ncomp = 1
cadet.root.input.model.unit_000.inlet_type = 'PIECEWISE_CUBIC_POLY'
cu = interpolate.CubicSpline(gauss_t, y)
n_sections=len(gauss_t)-1
for i in range(n_sections):
section_index = 'sec_{0:03d}'.format(i)
cadet.root.input.model.unit_000[section_index].cube_coeff = cu.c[:,i][0]
cadet.root.input.model.unit_000[section_index].qaud_coeff = cu.c[:,i][1]
cadet.root.input.model.unit_000[section_index].lin_coeff = cu.c[:,i][2]
cadet.root.input.model.unit_000[section_index].const_coeff = cu.c[:,i][3]
last_section_index = 'sec_{0:03d}'.format(n_sections)
cadet.root.input.model.unit_000[last_section_index].cube_coeff = [0.0]
cadet.root.input.model.unit_000[last_section_index].quad_coeff = [0.0]
cadet.root.input.model.unit_000[last_section_index].lin_coeff = [0.0]
cadet.root.input.model.unit_000[last_section_index].const_coeff = [0.0]
secs=gauss_t
secs=np.append(secs,t[-1])
cadet.root.input.solver.sections.section_times = secs # min
cadet.root.input.solver.sections.nsec = n_sections+1
cadet.root.input.solver.sections.section_continuity = [0,]
cadet.root.input.solver.user_solution_times = t
return cu
I basically define where the Gaussian should be placed and sections should be sliced and then append a last section with c=0 after that until the end of the solution time.
I call it with
t = np.linspace(0,1000,400) #The time at which the solution will be evaluated (start,end,steps/res)
gauss_t=np.linspace(0,200,100) #The time for the sections where the Gaussian will be placed (start,gauss_end,steps/res)
sigma=6.5 #sigma for Gaussian
t0=20 #mu for Gaussian
f=(1/sigma*np.sqrt(2*np.pi))*np.exp(-(gauss_t-t0)**2/(2*sigma)**2)
model = create_model_template(n_comp=1)
cu=get_spline_as_input(model, gauss_t, t, f)
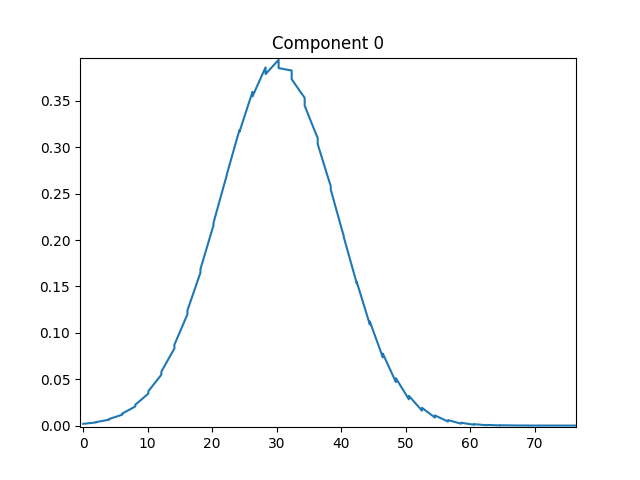
For small t0 values it works fine. But when I try to shift the pulse further to right (in time) by increasing t0, the simulation fails and gives
CompletedProcess(args=['C:/Users/Hannah Lanzrath/cadet/bin/cadet-cli', 'C:\\Users\\Hannah Lanzrath\\bsa7yvlv.h5'], returncode=3, stdout=b"[Error: idasErrorHandler::196] In function 'IDASolve' of module 'IDAS', error code 'IDA_ILL_INPUT':\r\nAt t = 70.7071, , mxstep steps taken before reaching tout.\r\n[Error: integrate::1355] IDASolve returned IDA_TOO_MUCH_WORK at t = 70.7071\r\n", stderr=b'SOLVER ERROR: Error in IDASolve: IDA_TOO_MUCH_WORK at t = 70.707071\r\n')
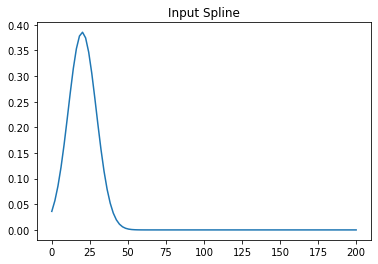
For example this input with t0=20 is working
but this input with t0=30 gives the error
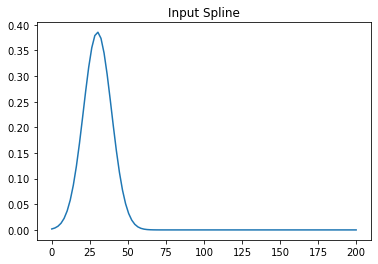
And I cannot really graps what could be the problem.
Cheers and thank you,
Hannah