Hello everyone,
Has anyone attempted to model the separation of peptides by reverse phase chromatography using CADET? What model is commonly used in CADET?I have chosen the Mobile Phase Modulator Langmuir (MPM) model, but currently I am struggling to find initial values for the model parameters. The parameters are incorrect and the calculation time is very long. Is there anyone who has developed a reverse phase chromatography model for peptide separation using CADET to communicate with?Or guide me on what to do next, thank you very much.
Hello everyone, when I am fitting the adsorption parameters. What is the reason for the continuous error message as shown below? How to make modifications?
Error:Evaluation of Comparator failed at [70.56860006 9.07499766 59.09685435 41.42399396] with Error “Simulation shape does not match experiment”. Returning bad metrics.
Hi chenkangnan,
Regarding your first question, unfortunately, I have no experience there, but hope one of the other users will comment on that soon.
Regarding your second post, if you have an issue with the CADET Code, please open an issue in the Troubleshooting section of this Forum. We are currently busy with the preparation of the CADET Workshop. I will come back to you ASAP, but if you still need help, it would be great if, in your new issue, you could share a minimal reproducible example (MRE) of your code that shows the issue that you encounter, including a comprehensive description of what you would expect for which command or method.
Kind regards,
Hannah
Thank you,h.lanzrath.I have resolved the second issue.
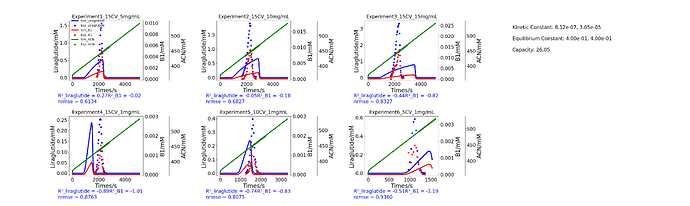
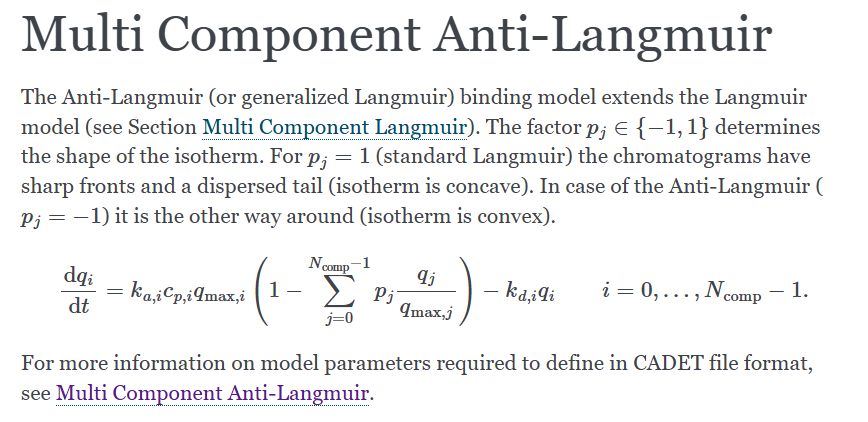
Hello everyone! I’m currently modeling the separation of liraglutide(a peptide) and impurities using reverse-phase chromatography. I conducted six different sets of calibration experiments.Experimental data reveals that under high sample loading conditions, the elution profile exhibits anti Langmuir behavior. Consequently, I intend to employ the anti-Langmuir model for modeling. However, this equation does not account for the influence of organic solvents. Therefore, I am doubtful about the applicability of this model to reverse-phase chromatography. Has anyone previously utilized models from Cadet for reverse-phase chromatography modeling, and if so, which model was typically employed.
Kindly help me with this.
Thanks
Chen
Hi Chen,
The first step is to understand how the solvent influences the parameters of the Langmuir model.
In CADET-Core, for stepwise isocratic processes, these parameters can be adjusted between time segments. For continuous solvent gradients, the parameters can be modified through externally specified functions.
In CADET-Process, the stepwise isocratic case is handled through events. Continuous solvent gradients are not yet supported in CADET-Process, but there is a workaround by exporting the model and applying the external function dependency in CADET-Core.
Eric
Hi lieres,
Thank you for your response.
I want to add an external function to AntiLangmuir to characterize the influence of organic solvents. According to this tutorial,I wrote
from CADETProcess.processModel import EXT_AntiLangmuir,
but I got an error:
ImportError: cannot import name 'EXT_AntiLangmuir' from 'CADETProcess.processModel' (E:\Anaconda\envs\ckn\Lib\site-packages\CADETProcess\processModel\__init__.py). Did you mean: 'AntiLangmuir'?
The name of the imported model is incorrect,how can I import it correctly?
Kindly help me with this.
Thanks
Chen
Hi lieres,
Am I supposed to modify the H5 file?
Yes,
external functions are currently not natively supported in CADET-Process. But since CADET-Process ultimately translates its config to the CADET-Core interface, you can modify it before running the simulation.
So you could to something like the following:
from CADETProcess.simulator import Cadet
simulator = Cadet()
cadet_config = simulator.get_process_config(process)
# Specify your external function here
from cadet import Cadet as CadetAPI
cadet = CadetAPI()
cadet.root = cadet_config
cadet.run_simulation()
Hi j.schmoelder,
Thank you for your response,but I’m still a bit confused about how to add additional functions specifically.![]()
This is my simulation code. The first component is acetonitrile, the second component is the target peptide, and the third component is an impurity. The model I have chosen is anti-Langmuir. I want to add an additional function (e.g. kd,i=kd,ads + kliner·Cacn) to the desorption constant to describe the influence of changes in acetonitrile concentration. Could you please advise me on how to Specify my external function?
import numpy as np
import matplotlib.pyplot as plt
from CADETProcess.processModel import ComponentSystem
from CADETProcess.processModel import AntiLangmuir
from CADETProcess.processModel import Inlet, LumpedRateModelWithoutPores, Outlet, Cstr
from CADETProcess.processModel import FlowSheet
from CADETProcess.processModel import Process
from CADETProcess.simulator import Cadet
from IPython.core.pylabtools import figsize
from sklearn.metrics import mean_squared_error
from scipy.interpolate import interp1d
from sklearn.metrics import r2_score
component_system = ComponentSystem()
component_system.add_component('Acetonitrile', molecular_weight=41.052)
component_system.add_component('Liraglutide', molecular_weight=3751.20)
component_system.add_component('B1', molecular_weight=3751.20)
EXPERIMENTS = [
{
'name': 'Experiment1_15CV_5mg/mL',
'file': 'experiment1_15CV_5mg.csv',
'length_CV': 15,
'load_duration_min': 2/0.7,
'feed_concentration': [50, 2.53825436114284, 0.0844733132291454*0.4*0.2]
},
{
'name': 'Experiment2_15CV_10mg/mL',
'file': 'experiment2_15CV_10mg.csv',
'length_CV': 15,
'load_duration_min': 2/0.7,
'feed_concentration': [50, 4.70906535527985, 0.175327739279275*0.4*0.2]
},
{
'name': 'Experiment3_15CV_15mg/mL',
'file': 'experiment3_15CV_15mg.csv',
'length_CV': 15,
'load_duration_min': 2/0.7,
'feed_concentration': [50, 7.49917863441825, 0.225849220442689*0.4*0.2]
},
{
'name': 'Experiment4_15CV_1mg/mL',
'file': 'experiment4_15CV_1mg.csv',
'length_CV': 15,
'load_duration_min': 0.5/0.7,
'feed_concentration': [50, 2.05457116438166, 0.0665269047490343*0.4*0.2]
},
{
'name': 'Experiment5_10CV_1mg/mL',
'file': 'experiment5_10CV_1mg.csv',
'length_CV': 10,
'load_duration_min': 0.5/0.7,
'feed_concentration': [50, 2.05457116438166, 0.0665269047490343*0.4*0.2]
},
]
experimental_data = []
for exp in EXPERIMENTS:
data = np.loadtxt(exp['file'], delimiter=',')
experimental_data.append({
'time': data[:, 0],
'ACN':data[:,1],
'liraglutide': data[:, 2],
'B1': data[:, 3],
'config': exp
})
def run_simulation(exp_config):
global adsorption_rate
global desorption_rate
global capacity
K_kin_liraglutide = 1.7642e1
K_kin_B1 = 5.2022e1
K_eq_liraglutide = 0.1236
K_eq_B1 = 0.1254
adsorption_rate = [0, K_eq_liraglutide / K_kin_liraglutide, K_eq_B1 / K_kin_B1] # (m3*m-3*s-1)
desorption_rate = [0, 1 / K_kin_liraglutide, 1 / K_kin_B1] # (s-1)
capacity = 100
binding_model = AntiLangmuir(component_system, name='AL')
binding_model.is_kinetic = True
binding_model.adsorption_rate = adsorption_rate
binding_model.desorption_rate = desorption_rate
# object.__setattr__(binding_model, 'desorption_rate_linear', [0.0, 0.1, 0.1])
binding_model.capacity = capacity
binding_model.antilangmuir = -1
Feed = Inlet(component_system, name='Feed')
Buffer = Inlet(component_system, name='Buffer')
Tube1 = LumpedRateModelWithoutPores(component_system, 'Tube1')
Tube1.total_porosity = 1
Tube1.length = 0.01 # m
Tube1.diameter = 0.00317 # m
Tube1.axial_dispersion = 1e-12
Tube1.c = [50, 0, 0]
Mixer = Cstr(component_system, 'Mixer')
Mixer.init_liquid_volume = 2.5e-8 # m3
Mixer.c = [50, 0, 0]
Tube2 = LumpedRateModelWithoutPores(component_system, 'Tube2')
Tube2.total_porosity = 1
Tube2.length = 0.4 # m
Tube2.diameter = 0.00025 # m
Tube2.axial_dispersion = 1e-12
Tube2.c = [50, 0, 0]
Value = Cstr(component_system, 'Value')
Value.init_liquid_volume = 5e-8 # m3
Value.c = [50, 0, 0]
Tube3 = LumpedRateModelWithoutPores(component_system, 'Tube3')
Tube3.total_porosity = 1
Tube3.length = 0.45 # m
Tube3.diameter = 0.00025 # m
Tube3.axial_dispersion = 1e-12
Tube3.c = [50, 0, 0]
Column = LumpedRateModelWithoutPores(component_system, name='Column')
Column.binding_model = binding_model
Column.total_porosity = 0.7062
Column.length = 0.25
Column.diameter = 0.0046
Column.axial_dispersion = 2.49e-8 # m2/s
Column.c = [50, 0, 0]
Column.q = [capacity, 0, 0]
Tube4 = LumpedRateModelWithoutPores(component_system, 'Tube4')
Tube4.total_porosity = 1
Tube4.length = 0.5 # m
Tube4.diameter = 0.00025 # m
Tube4.axial_dispersion = 1e-12 # m
Tube4.c = [50, 0, 0]
UV = Cstr(component_system, 'UV')
UV.init_liquid_volume = 1e-8 # m3
UV.c = [50, 0, 0]
Tube5 = LumpedRateModelWithoutPores(component_system, 'Tube5')
Tube5.total_porosity = 1
Tube5.length = 0.01 # m
Tube5.diameter = 0.00317 # m
Tube5.axial_dispersion = 1e-12 # m
Tube5.c = [50, 0, 0]
final_outlet = Outlet(component_system, name='final_outlet')
flow_sheet = FlowSheet(component_system)
flow_sheet.add_unit(Buffer)
flow_sheet.add_unit(Tube1)
flow_sheet.add_unit(Mixer)
flow_sheet.add_unit(Tube2)
flow_sheet.add_unit(Value)
flow_sheet.add_unit(Feed, feed_inlet=True)
flow_sheet.add_unit(Tube3)
flow_sheet.add_unit(Column)
flow_sheet.add_unit(Tube4)
flow_sheet.add_unit(UV)
flow_sheet.add_unit(Tube5)
flow_sheet.add_unit(final_outlet, product_outlet=True)
flow_sheet.add_connection(Buffer, Tube1)
flow_sheet.add_connection(Tube1, Mixer)
flow_sheet.add_connection(Mixer, Tube2)
flow_sheet.add_connection(Tube2, Value)
flow_sheet.add_connection(Feed, Value)
flow_sheet.add_connection(Value, Tube3)
flow_sheet.add_connection(Tube3, Column)
flow_sheet.add_connection(Column, Tube4)
flow_sheet.add_connection(Tube4, UV)
flow_sheet.add_connection(UV, Tube5)
flow_sheet.add_connection(Tube5, final_outlet)
CV = 0.0023 * 0.0023 * 3.14 * 0.25 # m3
length_CV = exp_config['length_CV']
load_duration_min = exp_config['load_duration_min']
Q = 0.7 / (60 * 1e6) # m3/s
t_load_end = 60 * load_duration_min
t_eq_end = 60 * 10
t_wash_start = 60 * 10
t_wash_duration = 60 * 10
t_wash_end = t_wash_start + t_wash_duration
t_elute_start = t_wash_end
t_elute_duration = length_CV * CV / Q
total_time = t_elute_start + t_elute_duration
process = Process(flow_sheet, exp_config['name'])
process.add_event('Load_on', 'flow_sheet.Feed.flow_rate', Q, time=0)
process.add_event('Buffer_off', 'flow_sheet.Buffer.flow_rate', 0, time=0)
process.add_event('Load_off', 'flow_sheet.Feed.flow_rate', 0, time=t_load_end)
process.add_event('Buffer_on', 'flow_sheet.Buffer.flow_rate', Q, time=t_load_end)
feed_conc = exp_config['feed_concentration']
process.add_event('Loading_on', 'flow_sheet.Feed.c',
[[feed_conc[0], 0, 0, 0], [feed_conc[1], 0, 0, 0], [feed_conc[2], 0, 0, 0]], time=0)
process.add_event('Buffering_off', 'flow_sheet.Buffer.c',
[[0, 0, 0, 0], [0, 0, 0, 0], [0, 0, 0, 0]], time=0)
process.add_event('Loading_off', 'flow_sheet.Feed.c',
[[0, 0, 0, 0], [0, 0, 0, 0], [0, 0, 0, 0]], time=t_load_end)
process.add_event('Equilibriuming_on', 'flow_sheet.Buffer.c',
[[50, 0, 0, 0], [0, 0, 0, 0], [0, 0, 0, 0]], time=t_load_end)
process.add_event('Washing_on', 'flow_sheet.Buffer.c',
[[50, (371.43 - 50) / t_wash_duration, 0, 0], [0, 0, 0, 0], [0, 0, 0, 0]],
time=t_wash_start)
process.add_event('Eluting_on', 'flow_sheet.Buffer.c',
[[371.43, (550 - 371.43) / t_elute_duration, 0, 0], [0, 0, 0, 0], [0, 0, 0, 0]],
time=t_elute_start)
process.cycle_time = total_time
# _ = process.plot_events()
simulator = Cadet()
simulator.timeout = 1
simulation_results = simulator.simulate(process)
return simulation_results
fig,axes = plt.subplots(2,3,figsize=(40,12))
plt.subplots_adjust(
left=0.1,
right=0.85,
bottom=0.15,
top=0.9,
wspace=1.0,
hspace=0.6
)
position = [[0,0],[0,1],[0,2],[1,0],[1,1],[1,2]]
for j in range(len(experimental_data)):
exp_data = experimental_data[j]
exp_config = exp_data['config']
simulation_results = run_simulation(exp_config)
sim_time = simulation_results.solution.final_outlet.outlet.time
sim_solution1 = simulation_results.solution.final_outlet.outlet.solution
sim_solution2 = simulation_results.solution.Column.outlet.solution
sim_liraglutide = sim_solution1[:, 1]
sim_B1 = sim_solution1[:, 2]
sim_ACN = sim_solution2[:, 0]
interp_liraglutide = interp1d(sim_time, sim_liraglutide, kind='linear',bounds_error=False, fill_value='extrapolate')
interp_B1 = interp1d(sim_time, sim_B1, kind='linear',bounds_error=False, fill_value='extrapolate')
interp_ACN = interp1d(sim_time, sim_ACN, kind='linear',bounds_error=False, fill_value='extrapolate')
exp_time = exp_data['time']
sim_liraglutide_interp = interp_liraglutide(exp_time)
sim_B1_interp = interp_B1(exp_time)
sim_ACN_interp = interp_ACN(exp_time)
print(j+1)
print(sim_liraglutide_interp)
R2_liraglutide = r2_score(exp_data['liraglutide'],sim_liraglutide_interp)
R2_B1 = r2_score(exp_data['B1'],sim_B1_interp)
s_liraglutide = np.max(exp_data['liraglutide']) - np.min(exp_data['liraglutide'])
s_B1 = np.max(exp_data['B1']) - np.min(exp_data['B1'])
nrmse_liraglutide = np.sqrt(mean_squared_error(exp_data['liraglutide'], sim_liraglutide_interp)) / s_liraglutide
nrmse_B1 = np.sqrt(mean_squared_error(exp_data['B1'], sim_B1_interp)) / s_B1
ax_primary = axes[position[j][0], position[j][1]]
ax_primary.plot(sim_time,sim_liraglutide, label='Sim_Liraglutide', color='blue')
ax_primary.scatter(exp_time, exp_data['liraglutide'],label='Exp_Liraglutide', color='blue', marker='o')
ax_primary.set_ylabel('Liraglutide/mM')
ax_primary.set_xlabel('Times/s')
ax_primary.set_title(exp_config['name'],pad = 10)
ax_primary.text(
-0.15, -0.22,
f"R²_liraglutide = {R2_liraglutide:.2f}"
f"R²_B1 = {R2_B1:.2f}\n"
f"nrmse = {nrmse_liraglutide + nrmse_B1 :.4f}",
transform=ax_primary.transAxes,
fontsize=20, color='blue',verticalalignment='top',horizontalalignment='left',
clip_on=False
)
ax_secondary = ax_primary.twinx()
ax_secondary.plot(sim_time, sim_B1, label='Sim_B1', color='red')
ax_secondary.scatter(exp_time, exp_data['B1'],
label='Exp_B1', color='red', marker='o')
ax_secondary.set_ylabel('B1/mM')
ax_secondary.set_ylim(top=(max(exp_data['B1'])*2))
ax_third = ax_primary.twinx()
ax_third.spines['right'].set_position(('outward', 110)) #向右移动距离
ax_third.plot(sim_time, sim_ACN, label='Sim_ACN', color='green')
ax_third.scatter(exp_time+250, exp_data['ACN'], label='Exp_ACN', marker='o',color='green')
ax_third.set_ylabel('ACN/mM')
fig.delaxes(axes[1,3])
plt.show()
Hi,
I think this would be a good topic for our next Office Hours. It’s easier to explain and give some hands-on support than to explain it here.
Hi j.schmoelder,
Thank you for your response. I know office hours is a great opportunity to clarify doubts, but my spoken English is relatively poor, so I may not be able to express my questions well. I will participate in office hours more often.Thank you again.