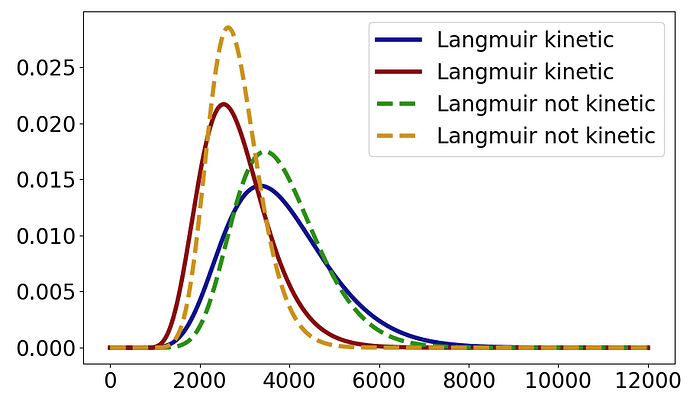
I do not understand the is_kinetic parameter. I got same results in CADET-Process for SMA binding using is_kinetic = True or False. Then, I tried reading Binding models — CADET and thought maybe it is only relevant for Langmuir and Langmuir LDF. But even then, the results for Langmuir with is_kinetic True is same as when is_kinetic is False.
In addition, the results for Langmuir LDF where I put driving_force_coefficient as 1 and appropriately adjust equilibrium_constant such that equilibrium_constant of LangmuirLDF is same as adorption/desorption of Langmuir, the results are again same and is_kinetic being True or False is not making any difference.
This is the code:
import numpy as np
from CADETProcess.processModel import ComponentSystem
from CADETProcess.processModel import Langmuir, LangmuirLDF
from CADETProcess.processModel import Inlet, GeneralRateModel, Outlet
from CADETProcess.processModel import FlowSheet
from CADETProcess.processModel import Process
component_system = ComponentSystem()
component_system.add_component('Protein 1')
component_system.add_component('Protein 2')
# binding_model = Langmuir(component_system, name='Lang')
# binding_model.is_kinetic = False
# binding_model.adsorption_rate = [1.3, 1.5]
# binding_model.desorption_rate = [0.5, 1.0]
# binding_model.capacity = 1
binding_model = LangmuirLDF(component_system, name='LangLDF')
binding_model.is_kinetic = True #False
binding_model.equilibrium_constant = [2.6, 1.5]
binding_model.driving_force_coefficient = [1.0, 1.0]
binding_model.capacity = 1
inlet = Inlet(component_system, name='inlet')
inlet.flow_rate = 2.88e-8
column = GeneralRateModel(component_system, name='column')
column.binding_model = binding_model
column.length = 0.65
column.diameter = 0.011286
column.bed_porosity = 0.37
column.particle_radius = 4.5e-5
column.particle_porosity = 0.33
column.axial_dispersion = 2.0e-7
column.film_diffusion = [2.0e-7, 2.0e-7]
column.pore_diffusion = [1e-7, 1e-7]
column.surface_diffusion = [0.0, 0.0]
outlet = Outlet(component_system, name='outlet')
# Flow Sheet
flow_sheet = FlowSheet(component_system)
flow_sheet.add_unit(inlet)
flow_sheet.add_unit(column)
flow_sheet.add_unit(outlet)
flow_sheet.add_connection(inlet, column)
flow_sheet.add_connection(column, outlet)
# Process
process = Process(flow_sheet, 'lwe')
process.cycle_time = 12000.0
## Create Events and Durations
wash_start = 400
_ = process.add_event('load', 'flow_sheet.inlet.c', [0.1, 0.1])
_ = process.add_event('wash', 'flow_sheet.inlet.c', [0.0, 0.0], wash_start)
column.c = [0, 0]
column.q = [0, 0]