Hi,
I am working on modeling Protein A affinity PCC chromatography. Since I am not very familiar with SMB Builder, I tried using the scheduling method.
Expected result:
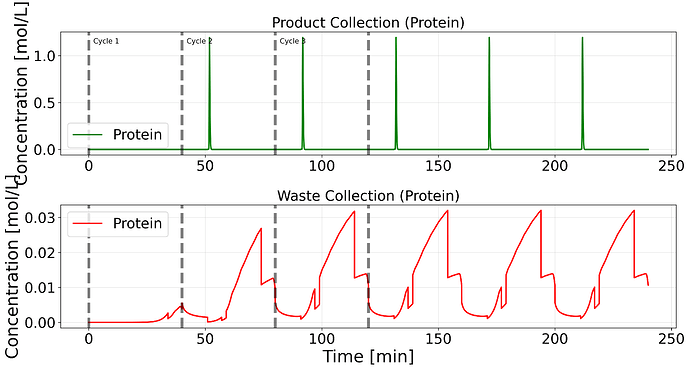
I was expecting a proper elution for different cycles.
Output:
Minimum reproducible code:
from CADETProcess.processModel import ComponentSystem, Inlet, Outlet, GeneralRateModel
from CADETProcess.processModel import FlowSheet, Process
from CADETProcess.processModel import MobilePhaseModulator
from CADETProcess.simulator import Cadet
import numpy as np
import matplotlib.pyplot as plt
def create_pcc_process():
CYCLE_TIME = 40 * 60
load_time = 37 * 60 # 37 minutes
wash_zone1_time = 3 * 60 # 3 minutes
# Zone 3 timings - total 40 minutes
t_w1 = 1 * 60 # 1 minute
t_w2 = 4 * 60 # 4 minutes
t_w3 = 6 * 60 # 6 minutes
t_elute = 6 * 60 # 6 minutes
t_clean1 = 2 * 60 # 2 minutes
t_pause = 15 * 60 # 15 minutes
t_clean2 = 1 * 60 # 1 minute
t_equil = 5 * 60 # 5 minutes
mLmin_to_m3s = 60 * 1e6
FLOW_LOAD = 3.03 / mLmin_to_m3s
FLOW_OTHER = 4.55 / mLmin_to_m3s
# pH and concentration parameters
mol_wt = 146.5
conc = 5.3
conc_load = conc / mol_wt # mol/L
pH_bind = 7
pH_elute = 3.5
pH_b_H = 10**(-pH_bind)
pH_e_H = 10**(-pH_elute)
#components
sys = ComponentSystem()
sys.add_component('Salt')
sys.add_component('Protein')
#binding model
binding_model = MobilePhaseModulator(sys, name='MPM')
binding_model.is_kinetic = False
binding_model.adsorption_rate = [0, 5.19e-08]
binding_model.desorption_rate = [0, 1.0]
binding_model.capacity = [0, 10]
binding_model.ion_exchange_characteristic = [0, 1.54]
binding_model.hydrophobicity = [0, 0]
binding_model.bound_states = [0, 1]
#Inlets
feed = Inlet(sys, 'feed')
feed.c = [pH_b_H, conc_load]
wash_z1 = Inlet(sys, 'wash_z1')
wash_z1.c = [pH_b_H, 0]
wash1 = Inlet(sys, 'wash1')
wash1.c = [pH_b_H, 0]
wash2 = Inlet(sys, 'wash2')
wash2.c = [pH_b_H, 0]
wash3 = Inlet(sys, 'wash3')
wash3.c = [pH_b_H, 0]
eluent = Inlet(sys, 'eluent')
eluent.c = [pH_e_H, 0]
cip = Inlet(sys, 'cip')
cip.c = [10**(-3.0), 0]
equil = Inlet(sys, 'equil')
equil.c = [pH_b_H, 0]
#columns
cols = []
for i in range(3):
column = GeneralRateModel(sys, name=f'col_{i+1}')
column.length = 0.058 # m
column.diameter = 0.01 # m
column.bed_porosity = 0.899998
column.particle_porosity = 0.49956
column.particle_radius = 50e-6
column.axial_dispersion = 9.21e-7
column.film_diffusion = [1e-4, 9.93e-5]
column.pore_diffusion = [1e-9, 9.54e-11]
column.binding_model = binding_model
column.discretization.ncol = 50
column.discretization.npar = 12
column.c = [pH_b_H, 0]
cols.append(column)
#Outlets
product = Outlet(sys, 'product')
waste = Outlet(sys, 'waste')
#flowsheet
fs = FlowSheet(sys)
for u in [feed, wash_z1, wash1, wash2, wash3, eluent, cip, equil, product, waste]:
fs.add_unit(u)
for c in cols:
fs.add_unit(c)
#connections
# Connect ALL inlets to ALL columns
for inlet in [feed, wash_z1, wash1, wash2, wash3, eluent, cip, equil]:
fs.add_connection(inlet, cols[0])
fs.add_connection(inlet, cols[1])
fs.add_connection(inlet, cols[2])
# Col1 output states: [product, waste, col_2]
fs.add_connection(cols[0], product)
fs.add_connection(cols[0], waste)
fs.add_connection(cols[0], cols[1])
# Col2 output states: [product, waste, col_3]
fs.add_connection(cols[1], product)
fs.add_connection(cols[1], waste)
fs.add_connection(cols[1], cols[2])
# Col3 output states: [product, waste, col_1]
fs.add_connection(cols[2], product)
fs.add_connection(cols[2], waste)
fs.add_connection(cols[2], cols[0])
#Process
process = Process(fs, 'PCC_Chromatography')
#total process time
process.cycle_time = 3 * CYCLE_TIME
#add events for all 3 phases
def add_phase_events(state_idx):
"""
state 0: Load C1->C2, Recover C3 (0-40 mins)
state 1: Load C2->C3, Recover C1 (40-80 mins)
state 2: Load C3->C1, Recover C2 (80-120 mins)
"""
t_base = state_idx * CYCLE_TIME
#[to_column1, to_column2, to_column3]
to_c1 = [1, 0, 0]
to_c2 = [0, 1, 0]
to_c3 = [0, 0, 1]
if state_idx == 0:
load_dest = to_c1
rec_inlet_map = to_c3
rec_col_name = 'col_3'
elif state_idx == 1:
load_dest = to_c2
rec_inlet_map = to_c1
rec_col_name = 'col_1'
else:
load_dest = to_c3
rec_inlet_map = to_c2
rec_col_name = 'col_2'
#LOADING (0-37 min): Z1
process.add_event(f'S{state_idx}_load_start', 'flow_sheet.feed.flow_rate', FLOW_LOAD, t_base)
process.add_event(f'S{state_idx}_load_dest', 'flow_sheet.output_states.feed', load_dest, t_base)
if state_idx == 0:
process.add_event(f'S{state_idx}_c1_conn', 'flow_sheet.output_states.col_1', [0, 0, 1], t_base) # C1->C2
process.add_event(f'S{state_idx}_c2_conn', 'flow_sheet.output_states.col_2', [0, 1, 0], t_base) # C2->Waste
elif state_idx == 1:
process.add_event(f'S{state_idx}_c2_conn', 'flow_sheet.output_states.col_2', [0, 0, 1], t_base) # C2->C3
process.add_event(f'S{state_idx}_c3_conn', 'flow_sheet.output_states.col_3', [0, 1, 0], t_base) # C3->Waste
else:
process.add_event(f'S{state_idx}_c3_conn', 'flow_sheet.output_states.col_3', [0, 0, 1], t_base) # C3->C1
process.add_event(f'S{state_idx}_c1_conn', 'flow_sheet.output_states.col_1', [0, 1, 0], t_base) # C1->Waste
#WASH (37-40 min): Z1
t_chase = t_base + load_time
process.add_event(f'S{state_idx}_load_stop', 'flow_sheet.feed.flow_rate', 0.0, t_chase)
process.add_event(f'S{state_idx}_z1w_start', 'flow_sheet.wash_z1.flow_rate', FLOW_LOAD, t_chase)
process.add_event(f'S{state_idx}_z1w_dest', 'flow_sheet.output_states.wash_z1', load_dest, t_chase)
# Stop
process.add_event(f'S{state_idx}_z1w_stop', 'flow_sheet.wash_z1.flow_rate', 0.0, t_base + CYCLE_TIME)
#Z2: RECOVERY STEPS
cur_t = t_base
def set_rec_out(time, dest_type):
vec = [0, 1, 0] if dest_type == 'waste' else [1, 0, 0]
process.add_event(f'{rec_col_name}_out_{state_idx}_{int(time)}',
f'flow_sheet.output_states.{rec_col_name}', vec, time)
#Wash 1
process.add_event(f'w1_start_{state_idx}', 'flow_sheet.wash1.flow_rate', FLOW_OTHER, cur_t)
process.add_event(f'w1_dest_{state_idx}', 'flow_sheet.output_states.wash1', rec_inlet_map, cur_t)
set_rec_out(cur_t, 'waste')
cur_t += t_w1
process.add_event(f'w1_stop_{state_idx}', 'flow_sheet.wash1.flow_rate', 0.0, cur_t)
#Wash 2
process.add_event(f'w2_start_{state_idx}', 'flow_sheet.wash2.flow_rate', FLOW_OTHER, cur_t)
process.add_event(f'w2_dest_{state_idx}', 'flow_sheet.output_states.wash2', rec_inlet_map, cur_t)
cur_t += t_w2
process.add_event(f'w2_stop_{state_idx}', 'flow_sheet.wash2.flow_rate', 0.0, cur_t)
#Wash 3
process.add_event(f'w3_start_{state_idx}', 'flow_sheet.wash3.flow_rate', FLOW_OTHER, cur_t)
process.add_event(f'w3_dest_{state_idx}', 'flow_sheet.output_states.wash3', rec_inlet_map, cur_t)
cur_t += t_w3
process.add_event(f'w3_stop_{state_idx}', 'flow_sheet.wash3.flow_rate', 0.0, cur_t)
#Elution
process.add_event(f'elute_start_{state_idx}', 'flow_sheet.eluent.flow_rate', FLOW_OTHER, cur_t)
process.add_event(f'elute_dest_{state_idx}', 'flow_sheet.output_states.eluent', rec_inlet_map, cur_t)
set_rec_out(cur_t, 'product')
cur_t += t_elute
process.add_event(f'elute_stop_{state_idx}', 'flow_sheet.eluent.flow_rate', 0.0, cur_t)
#Clean 1
process.add_event(f'cip1_start_{state_idx}', 'flow_sheet.cip.flow_rate', FLOW_OTHER, cur_t)
process.add_event(f'cip1_dest_{state_idx}', 'flow_sheet.output_states.cip', rec_inlet_map, cur_t)
set_rec_out(cur_t, 'waste')
cur_t += t_clean1
#Pause
process.add_event(f'pause_start_{state_idx}', 'flow_sheet.cip.flow_rate', 0.0, cur_t)
cur_t += t_pause
#Clean 2
process.add_event(f'cip2_start_{state_idx}', 'flow_sheet.cip.flow_rate', FLOW_OTHER, cur_t)
cur_t += t_clean2
process.add_event(f'cip2_stop_{state_idx}', 'flow_sheet.cip.flow_rate', 0.0, cur_t)
#Equilibration
process.add_event(f'eq_start_{state_idx}', 'flow_sheet.equil.flow_rate', FLOW_OTHER, cur_t)
process.add_event(f'eq_dest_{state_idx}', 'flow_sheet.output_states.equil', rec_inlet_map, cur_t)
cur_t += t_equil
process.add_event(f'eq_stop_{state_idx}', 'flow_sheet.equil.flow_rate', 0.0, cur_t)
# Add events
add_phase_events(0)
add_phase_events(1)
add_phase_events(2)
print(f"\nProcess created with {len(process.events_dict)} events")
return process
def run_pcc_simulation():
process = create_pcc_process()
simulator = Cadet()
simulator.evaluate_stationarity = True
simulator.n_cycles_max = 1
simulator.n_cycles_min = 1
simulator.timeout = 3600 # 1 hour timeout
try:
simulation_results = simulator.simulate(process)
print(f"Simulation completed successfully!")
print(f"Cycles simulated: {simulation_results.n_cycles}")
return process, simulation_results
except Exception as e:
print(f"Simulation failed with error: {e}")
import traceback
traceback.print_exc()
return process, None
def plot_results(process, simulation_results):
if simulation_results is None:
print("No results to plot")
return
try:
fig, axes = plt.subplots(2, 1, figsize=(14, 8))
fig.suptitle('PCC Chrom Process', fontsize=16)
# Plot Product outlet
ax = axes[0]
if hasattr(simulation_results.solution, 'product'):
product_data = simulation_results.solution.product.outlet.solution
product_time = simulation_results.solution.product.outlet.time / 60
ax.plot(product_time, product_data[:, 1], label='Protein', color='green', linewidth=2)
# Mark cycle switches
for i in range(4):
cycle_start = i * 40
ax.axvline(x=cycle_start, color='k', linestyle='--', alpha=0.5)
if i < 3:
ax.text(cycle_start + 2, ax.get_ylim()[1]*0.9, f'Cycle {i+1}', fontsize=10)
ax.set_ylabel('Concentration [mol/L]')
ax.set_title('Product Collection (Protein)')
ax.legend()
ax.grid(True, alpha=0.3)
# Plot Waste outlet
ax = axes[1]
if hasattr(simulation_results.solution, 'waste'):
waste_data = simulation_results.solution.waste.outlet.solution
waste_time = simulation_results.solution.waste.outlet.time / 60
ax.plot(waste_time, waste_data[:, 1], label='Protein', color='red', linewidth=2)
# Mark cycle switches
for i in range(4):
cycle_start = i * 40
ax.axvline(x=cycle_start, color='k', linestyle='--', alpha=0.5)
ax.set_xlabel('Time [min]')
ax.set_ylabel('Concentration [mol/L]')
ax.set_title('Waste Collection (Protein)')
ax.legend()
ax.grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('pcc_chromatography_results.png', dpi=300, bbox_inches='tight')
print("\nResults saved as: pcc_chromatography_results.png")
plt.show()
except Exception as e:
print(f"Error plotting results: {e}")
import traceback
traceback.print_exc()
if __name__ == "__main__":
process, results = run_pcc_simulation()
if results:
plot_results(process, results)
else:
print("\nSimulation failed")
Kindly help me with this.
Thanks
Naveen J