Hi There,
I am currently trying to generate a reference simulation for the Multi-State SMA isotherm with 2 components ‘salt’, and ‘A’ in CADET-Process.
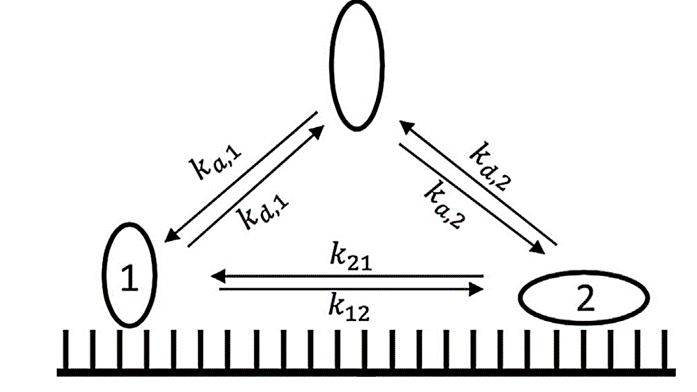
I have set the number of bound states for component ‘A’ to 2 and for salt to 1 as required by CADET. I notice that for adsorption and desorption rates, 3 inputs are therefore required to account for one ‘salt = 0’ and the two states of component ‘A’. However when defining ‘conversion_rate’ between bound states CADET asks for 4 inputs. I would have thought a ‘0’ input would be required for salt then two values for the forward and backward conversion between states giving 3 again. Where does the extra number come from and what is the structure of this input?
I think I must be misunderstanding something as when running the below code I get the following error of:
CADETProcessError: CADET Error: Simulation failed with b’ERROR: Not enough elements in dataset MSSMA_RATES (expected at least 5 but got only 4)\r\n’
Many Thanks for your help.
import numpy as np
from CADETProcess.processModel import ComponentSystem
from CADETProcess.processModel import MultistateStericMassAction
from CADETProcess.processModel import Inlet, GeneralRateModel, Outlet
from CADETProcess.processModel import FlowSheet
from CADETProcess.processModel import Process
component_system = ComponentSystem()
component_system.add_component('Salt')
component_system.add_component('A')
binding_model = MultistateStericMassAction(component_system,bound_states=[1,2], name='MSSMA')
binding_model.is_kinetic = True
binding_model.adsorption_rate = [0.0, 35.5,60]
binding_model.desorption_rate = [0.0, 1000,1000]
binding_model.conversion_rate = [0,0,20,70]
binding_model.characteristic_charge = [0.0, 4.7,10]
binding_model.steric_factor = [0.0, 11.83,20]
binding_model.capacity = 1200.0
inlet = Inlet(component_system, name='inlet')
inlet.flow_rate = 6.683738370512285e-8
column = GeneralRateModel(component_system, name='column')
column.binding_model = binding_model
column.length = 0.014
column.diameter = 0.02
column.bed_porosity = 0.37
column.particle_radius = 4.5e-5
column.particle_porosity = 0.75
column.axial_dispersion = 5.75e-8
column.film_diffusion = column.n_comp*[6.9e-6]
column.pore_diffusion = [7e-10, 6.07e-11]
column.surface_diffusion = column.n_bound_states*[0.0]
column.c = [50, 0]
column.cp = [50, 0]
column.q = [binding_model.capacity, 0,0]
outlet = Outlet(component_system, name='outlet')
flow_sheet = FlowSheet(component_system)
flow_sheet.add_unit(inlet)
flow_sheet.add_unit(column)
flow_sheet.add_unit(outlet, product_outlet=True)
flow_sheet.add_connection(inlet, column)
flow_sheet.add_connection(column, outlet)
process = Process(flow_sheet, 'lwe')
process.cycle_time = 2000.0
load_duration = 9
t_gradient_start = 90.0
gradient_duration = process.cycle_time - t_gradient_start
c_load = np.array([50.0, 1.0])
c_wash = np.array([50.0, 0.0])
c_elute = np.array([500.0, 0.0])
gradient_slope = (c_elute - c_wash)/gradient_duration
c_gradient_poly = np.array(list(zip(c_wash, gradient_slope)))
process.add_event('load', 'flow_sheet.inlet.c', c_load)
process.add_event('wash', 'flow_sheet.inlet.c', c_wash, load_duration)
process.add_event('grad_start', 'flow_sheet.inlet.c', c_gradient_poly, t_gradient_start)
if __name__ == '__main__':
from CADETProcess.simulator import Cadet
process_simulator = Cadet()
simulation_results = process_simulator.simulate(process)
from CADETProcess.plotting import SecondaryAxis
sec = SecondaryAxis()
sec.components = ['Salt']
sec.y_label = '$c_{salt}$'
simulation_results.solution.column.outlet.plot(secondary_axis=sec)