Hey ![]() as the title suggest i have run into a problem when trying to simulate my model.
as the title suggest i have run into a problem when trying to simulate my model.
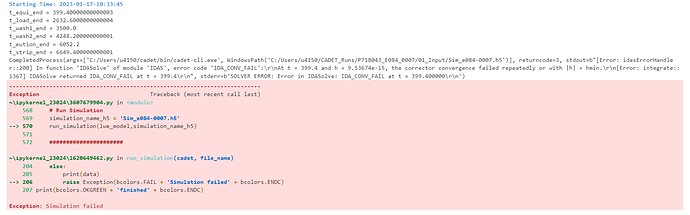
The Simulation works fine when running it in quasi-stationary binding mode. But when I switch over to a kinetic binding mode the following error occurs.
The Problem seems to be at a section switch, but I don’t know how to solve it. I guess it has something to do with the complexity of the SMA model, since the error only occurred after adjusting the characteristic charge to 68 ( before it was around 4 and the simulation worked fine).
I would appreciate any help.
Greetings
David
Code :
# Timestamp creation
now = datetime.now()
date_start_0 = datetime.date(now)
hour_start_0 = now.hour
minute_start_0 = now.minute
second_start_0 = now.second
timestamp_start_0 = str(date_start_0)+'-'+str(hour_start_0)+'-'+str(minute_start_0)+'-'+str(second_start_0)
time_start_0 = str(date_start_0)+'-'+str(hour_start_0)+':'+str(minute_start_0)+':'+str(second_start_0)
print(bcolors.OKBLUE + 'Starting Time: ' + time_start_0 + bcolors.ENDC)
## Reading options
input_reading = False # True for input reading, False for manual input parameters (below)
input_file = 'input'
run_name = "P718043_E084_0007"
## Saving options
saving_csv = False # True for saving the csv, False for non saving
saving_plot = False # True for saving the plot, False for non saving
combining_output_data = True
start_row = 0
row = start_row
stop_row = 84
while row < stop_row:
# Timestamp creation
now = datetime.now()
date_start = datetime.date(now)
hour_start = now.hour
minute_start = now.minute
second_start = now.second
# Names of saved data
combined_data_filename = timestamp_start_0 + 'combined.csv'
Operating_Person = 'David Achauer' # 'shorthand symbol and first name + last name in brackets'
Simulation_name_0 = 'Master'
if input_reading:
Simulation_name = Simulation_name_0 +'_row'+str(row) # 'Desired name for the exported file'
else:
Simulation_name = Simulation_name_0 # 'Desired name for the exported file'
## Operating Parameters
# Column parameters
col_poros = 0.7
length_0 = 12.01 # [m]
length_1 = 1.5# [m]
length_2 = 0.0005/(1-col_poros) # [m] unit_009
length_membran = 0.108 # [m]
lengthcomb = length_1+length_membran
column_diameter = 0.0225 # [m]
A_column = (column_diameter ** 2)*np.pi/4 # [m^2]
cv = A_column * length_2 # [m^3]
par_poros = 0.48
par_radius = 0.5e-5 # [m]
tube_diameter = 0.000762 # [m]
A_tube = np.pi*(tube_diameter**2)/4 # [m^2]
Q_mlmin = 3 # [ml/min]
Q = Q_mlmin / 60000000 # [m^3/s]
# Operation times +200e-6
volume_equi_end = 19.97e-6 # [m^3]
volume_load_end = 131.63e-6 # [m^3]
volume_wash1_end = 175.00e-6 # [m^3]
volume_wash2_end = 212.41e-6
volume_elution_end = 302.61e-6
volume_strip_end = 332.47e-6
t_equi_end = volume_equi_end/Q
t_load_end = volume_load_end/Q
t_wash1_end = volume_wash1_end/Q
t_wash2_end = volume_wash2_end/Q
t_elution_end = volume_elution_end/Q
t_strip_end = volume_strip_end/Q
t_end = t_strip_end
print('t_equi_end =',t_equi_end)
print('t_load_end =',t_load_end)
print('t_wash1_end =',t_wash1_end)
print('t_wash2_end =',t_wash2_end)
print('t_eution_end =',t_elution_end)
print('t_strip_end =',t_end)
# Concentrations of the relevant components
# pDNA experiments
c_pDNA = 0.000113333
c_salt_equi = 610
c_init = [c_salt_equi, 0.0]
c_salt_load = 610
c_salt_wash1 = 610
c_salt_wash2 = 630
c_salt_elution = 750
c_salt_strip = 1000
# Number of components
n_comp = len(c_init)
## Determined Parameters
# CSTRS
V_CSTR_0 = 2.22E-06 # [m^3]
V_CSTR_1 = 8.37E-08 # [m^3]
V_CSTR_2 = 1.33E-07# [m^3]
V_CSTR_Membran = 1.69E-09 #[m^3]
# Parameter for Mass Transfer and Dispersion
col_disp_0 = 6.55E-07 # [m^2/s]
col_disp_1 = 2.99E-08 # [m^2/s]
col_disp_2 = 1.48E-08 # [m^2/s]
col_disp_Membran = 0.000365 #[m^2/s]
film_diff_2 = [1,1] # [m/s]
par_diff_2 = [1, 1] # [m^2/s]
surf_diff_2 = [0, 0] # [m^2/s]
# Parameters for Steric Mass Action
ka = [0, 4.76e7] #2e41
kd = [0, 1]
Nu = [0, 68.4]
sigma = [0,60]
lambda_ = 530.87
q_init = [lambda_, 0]
# parameters for discretization
n_column = 30
n_part = 30
row = stop_row + 1 # so the while loop gets interrupted after one cycle
# Number of units
lwe_model = get_cadet_template(n_units=19)
#### pDNA Run ####
## Sections
lwe_model.root.input.solver.sections.nsec = 6
lwe_model.root.input.solver.sections.section_times = [0.0,t_equi_end,t_load_end,t_wash1_end,t_wash2_end,t_elution_end,t_strip_end] # s
# Equi
lwe_model.root.input.model.unit_000.sec_000.const_coeff = [c_salt_equi,0]# [mM]
lwe_model.root.input.model.unit_003.sec_000.const_coeff = [c_salt_equi,0] # [mM]
# Load
lwe_model.root.input.model.unit_000.sec_001.const_coeff = [c_salt_load,0]# [mM]
lwe_model.root.input.model.unit_003.sec_001.const_coeff = [c_salt_load,c_pDNA] # [mM]
# Wash1
lwe_model.root.input.model.unit_000.sec_002.const_coeff = [c_salt_wash1,0] # [mM]
lwe_model.root.input.model.unit_003.sec_002.const_coeff = [c_salt_wash1,0] # [mM]
# Wash2
lwe_model.root.input.model.unit_000.sec_003.const_coeff = [c_salt_wash2,0] # [mM]
lwe_model.root.input.model.unit_003.sec_003.const_coeff = [c_salt_wash2,0] # [mM]
#Elution
lwe_model.root.input.model.unit_000.sec_004.const_coeff = [c_salt_elution,0] # [mM]
lwe_model.root.input.model.unit_003.sec_004.const_coeff = [c_salt_elution,0] # [mM]
# Strip
lwe_model.root.input.model.unit_000.sec_005.const_coeff = [c_salt_strip,0] # [mM]
lwe_model.root.input.model.unit_003.sec_005.const_coeff = [c_salt_strip,0] # [mM]
# Set the times that the simulator writes out data for
lwe_model.root.input.solver.user_solution_times = np.linspace(0, t_end, 2*(int(t_end)+1))
## Units
# INLET - Buffer
lwe_model.root.input.model.unit_000.unit_type = 'INLET'
lwe_model.root.input.model.unit_000.ncomp = n_comp
lwe_model.root.input.model.unit_000.inlet_type = 'PIECEWISE_CUBIC_POLY'
# CSTR_0
lwe_model.root.input.model.unit_001.unit_type = 'CSTR'
lwe_model.root.input.model.unit_001.ncomp = n_comp
lwe_model.root.input.model.unit_001.init_volume = V_CSTR_0 # [m^3]
lwe_model.root.input.model.unit_001.init_c = c_init # [mM]
# Tubular reactor
lwe_model.root.input.model.unit_002.unit_type = 'LUMPED_RATE_MODEL_WITHOUT_PORES'
lwe_model.root.input.model.unit_002.ncomp = n_comp
lwe_model.root.input.model.unit_002.col_length = length_0 # [m]
lwe_model.root.input.model.unit_002.cross_section_area = A_tube # [m^2]
lwe_model.root.input.model.unit_002.total_porosity = 1
lwe_model.root.input.model.unit_002.col_dispersion = col_disp_0 # [m^2/s]
lwe_model.root.input.model.unit_002.init_c = c_init # [mM]
lwe_model.root.input.model.unit_002.init_q = n_comp*[0.0] # [mM]
lwe_model.root.input.model.unit_002.adsorption_model = 'NONE'
# INLET - Inlet
lwe_model.root.input.model.unit_003.unit_type = 'INLET'
lwe_model.root.input.model.unit_003.ncomp = n_comp
lwe_model.root.input.model.unit_003.inlet_type = 'PIECEWISE_CUBIC_POLY'
# CSTR_1
lwe_model.root.input.model.unit_004.unit_type = 'CSTR'
lwe_model.root.input.model.unit_004.ncomp = n_comp
lwe_model.root.input.model.unit_004.init_volume = V_CSTR_1 # [m^3]
lwe_model.root.input.model.unit_004.init_c = c_init # [mM]
# Tubular reactor
lwe_model.root.input.model.unit_005.unit_type = 'LUMPED_RATE_MODEL_WITHOUT_PORES'
lwe_model.root.input.model.unit_005.ncomp = n_comp
lwe_model.root.input.model.unit_005.col_length = lengthcomb # [m]
lwe_model.root.input.model.unit_005.cross_section_area = A_tube # [m^2]
lwe_model.root.input.model.unit_005.total_porosity = 1
lwe_model.root.input.model.unit_005.col_dispersion = col_disp_1 # [m^2/s]
lwe_model.root.input.model.unit_005.init_c = c_init # [mM]
lwe_model.root.input.model.unit_005.init_q = n_comp*[0.0] # [mM]
lwe_model.root.input.model.unit_005.adsorption_model = 'NONE'
# General rate model + Steric_mass_action
#we_model.root.input.model.unit_006.unit_type = 'GENERAL_RATE_MODEL'
lwe_model.root.input.model.unit_006.unit_type = 'LUMPED_RATE_MODEL_WITH_PORES'
lwe_model.root.input.model.unit_006.ncomp = n_comp
lwe_model.root.input.model.unit_006.col_length = length_2 # [m]
lwe_model.root.input.model.unit_006.cross_section_area = A_column # [m^2]
lwe_model.root.input.model.unit_006.col_porosity = col_poros
lwe_model.root.input.model.unit_006.par_porosity = par_poros
lwe_model.root.input.model.unit_006.par_radius = par_radius # [m]
lwe_model.root.input.model.unit_006.col_dispersion = col_disp_2 # [m^2/s]
lwe_model.root.input.model.unit_006.film_diffusion = film_diff_2 # [m/s7
lwe_model.root.input.model.unit_006.par_diffusion = par_diff_2 # [m^2/s]
lwe_model.root.input.model.unit_006.par_surfdiffusion = surf_diff_2 # [m^2/s]
lwe_model.root.input.model.unit_006.init_c = c_init # [mM]
lwe_model.root.input.model.unit_006.init_q = q_init # [mM]
lwe_model.root.input.model.unit_006.adsorption_model = 'STERIC_MASS_ACTION'
lwe_model.root.input.model.unit_006.adsorption.is_kinetic = 1
lwe_model.root.input.model.unit_006.adsorption.sma_ka = ka # [m^3/s*m^3]
lwe_model.root.input.model.unit_006.adsorption.sma_kd = kd # [1/s]
lwe_model.root.input.model.unit_006.adsorption.sma_lambda = lambda_ # [mol/m^3]
lwe_model.root.input.model.unit_006.adsorption.sma_nu = Nu
lwe_model.root.input.model.unit_006.adsorption.sma_sigma = sigma
#lwe_model.root.input.model.unit_006.adsorption.sma_refq = lambda_ # [mol*m^3]
#lwe_model.root.input.model.unit_006.adsorption.sma_refc0 = c_salt_strip
lwe_model.root.input.model.unit_013.unit_type = 'LUMPED_RATE_MODEL_WITH_PORES'
lwe_model.root.input.model.unit_013.ncomp = n_comp
lwe_model.root.input.model.unit_013.col_length = length_2 # [m]
lwe_model.root.input.model.unit_013.cross_section_area = A_column # [m^2]
lwe_model.root.input.model.unit_013.col_porosity = col_poros
lwe_model.root.input.model.unit_013.par_porosity = par_poros
lwe_model.root.input.model.unit_013.par_radius = par_radius # [m]
lwe_model.root.input.model.unit_013.col_dispersion = col_disp_2 # [m^2/s]
lwe_model.root.input.model.unit_013.film_diffusion = film_diff_2 # [m/s7
lwe_model.root.input.model.unit_013.par_diffusion = par_diff_2 # [m^2/s]
lwe_model.root.input.model.unit_013.par_surfdiffusion = surf_diff_2 # [m^2/s]
lwe_model.root.input.model.unit_013.init_c = c_init # [mM]
lwe_model.root.input.model.unit_013.init_q = q_init # [mM]
lwe_model.root.input.model.unit_013.adsorption_model = 'STERIC_MASS_ACTION'
lwe_model.root.input.model.unit_013.adsorption.is_kinetic = 1
lwe_model.root.input.model.unit_013.adsorption.sma_ka = ka # [m^3/s*m^3]
lwe_model.root.input.model.unit_013.adsorption.sma_kd = kd # [1/s]
lwe_model.root.input.model.unit_013.adsorption.sma_lambda = lambda_ # [mol/m^3]
lwe_model.root.input.model.unit_013.adsorption.sma_nu = Nu
lwe_model.root.input.model.unit_013.adsorption.sma_sigma = sigma
## Connections
Ant1 = 0.3
Ant2 = 0.3
Ant3 = 0.2
Ant4 = 0.15
Ant5 = 0.05
Vtail1 = 4e-8
Vtail2 = 3.55E-07
Vtail3 = 2.98E-07
Vtail4 = 1.74E-07
Vtail5 = 1.22E-07
# Zonal Rate Model
lwe_model.root.input.model.unit_007.unit_type = 'CSTR'
lwe_model.root.input.model.unit_007.ncomp = n_comp
lwe_model.root.input.model.unit_007.init_volume = Vtail1 # [m^3]
lwe_model.root.input.model.unit_007.init_c = c_init # [mM]
lwe_model.root.input.model.unit_008.unit_type = 'CSTR'
lwe_model.root.input.model.unit_008.ncomp = n_comp
lwe_model.root.input.model.unit_008.init_volume = Vtail2 # [m^3]
lwe_model.root.input.model.unit_008.init_c = c_init # [mM]
lwe_model.root.input.model.unit_009.unit_type = 'CSTR'
lwe_model.root.input.model.unit_009.ncomp = n_comp
lwe_model.root.input.model.unit_009.init_volume = Vtail3 # [m^3]
lwe_model.root.input.model.unit_009.init_c = c_init # [mM]
lwe_model.root.input.model.unit_011.unit_type = 'CSTR'
lwe_model.root.input.model.unit_011.ncomp = n_comp
lwe_model.root.input.model.unit_011.init_volume = Vtail4 # [m^3]
lwe_model.root.input.model.unit_011.init_c = c_init # [mM]
lwe_model.root.input.model.unit_012.unit_type = 'CSTR'
lwe_model.root.input.model.unit_012.ncomp = n_comp
lwe_model.root.input.model.unit_012.init_volume = Vtail5 # [m^3]
lwe_model.root.input.model.unit_012.init_c = c_init # [mM]
lwe_model.root.input.model.unit_014.unit_type = 'CSTR'
lwe_model.root.input.model.unit_014.ncomp = n_comp
lwe_model.root.input.model.unit_014.init_volume = Vtail1 # [m^3]
lwe_model.root.input.model.unit_014.init_c = c_init # [mM]
lwe_model.root.input.model.unit_015.unit_type = 'CSTR'
lwe_model.root.input.model.unit_015.ncomp = n_comp
lwe_model.root.input.model.unit_015.init_volume = Vtail2 # [m^3]
lwe_model.root.input.model.unit_015.init_c = c_init # [mM]
lwe_model.root.input.model.unit_016.unit_type = 'CSTR'
lwe_model.root.input.model.unit_016.ncomp = n_comp
lwe_model.root.input.model.unit_016.init_volume = Vtail3 # [m^3]
lwe_model.root.input.model.unit_016.init_c = c_init # [mM]
lwe_model.root.input.model.unit_017.unit_type = 'CSTR'
lwe_model.root.input.model.unit_017.ncomp = n_comp
lwe_model.root.input.model.unit_017.init_volume = Vtail4 # [m^3]
lwe_model.root.input.model.unit_017.init_c = c_init # [mM]
lwe_model.root.input.model.unit_018.unit_type = 'CSTR'
lwe_model.root.input.model.unit_018.ncomp = n_comp
lwe_model.root.input.model.unit_018.init_volume = Vtail5 # [m^3]
lwe_model.root.input.model.unit_018.init_c = c_init # [mM]
# OUTLET
lwe_model.root.input.model.unit_010.unit_type = 'OUTLET'
lwe_model.root.input.model.unit_010.ncomp = n_comp
flow_changes = True
if flow_changes:
lwe_model.root.input.model.connections.connections_include_dynamic_flow = 0
lwe_model.root.input.model.connections.nswitches = 6
# Equi
lwe_model.root.input.model.connections.switch_000.section = 0
lwe_model.root.input.model.connections.switch_000.connections = [0, 1, -1, -1, Q,
1, 2, -1, -1, Q,
2, 4, -1, -1, Q,
3, 4, -1, -1, 0,
4, 5, -1, -1, Q,
5, 6, -1, -1, Q,
6, 7, -1, -1, Q*Ant1,
6, 8, -1, -1, Q*Ant2,
6, 9, -1, -1, Q*Ant3,
6, 11, -1, -1, Q*Ant4,
6, 12, -1, -1, Q*Ant5,
7, 13, -1, -1, Q*Ant1,
8, 13, -1, -1, Q*Ant2,
9, 13, -1, -1, Q*Ant3,
11, 13, -1, -1, Q*Ant4,
12, 13, -1, -1, Q*Ant5,
13, 14, -1, -1, Q*Ant1,
13, 15, -1, -1, Q*Ant2,
13, 16, -1, -1, Q*Ant3,
13, 17, -1, -1, Q*Ant4,
13, 18, -1, -1, Q*Ant5,
14, 10, -1, -1, Q*Ant1,
15, 10, -1, -1, Q*Ant2,
16, 10, -1, -1, Q*Ant3,
17, 10, -1, -1, Q*Ant4,
18, 10, -1, -1, Q*Ant5,
]
# Load
lwe_model.root.input.model.connections.switch_001.section = 1
lwe_model.root.input.model.connections.switch_001.connections = [0, 1, -1, -1, 0,
1, 2, -1, -1, 0,
2, 4, -1, -1, 0,
3, 4, -1, -1, Q,
4, 5, -1, -1, Q,
5, 6, -1, -1, Q,
6, 7, -1, -1, Q*Ant1,
6, 8, -1, -1, Q*Ant2,
6, 9, -1, -1, Q*Ant3,
6, 11, -1, -1, Q*Ant4,
6, 12, -1, -1, Q*Ant5,
7, 13, -1, -1, Q*Ant1,
8, 13, -1, -1, Q*Ant2,
9, 13, -1, -1, Q*Ant3,
11, 13, -1, -1, Q*Ant4,
12, 13, -1, -1, Q*Ant5,
13, 14, -1, -1, Q*Ant1,
13, 15, -1, -1, Q*Ant2,
13, 16, -1, -1, Q*Ant3,
13, 17, -1, -1, Q*Ant4,
13, 18, -1, -1, Q*Ant5,
14, 10, -1, -1, Q*Ant1,
15, 10, -1, -1, Q*Ant2,
16, 10, -1, -1, Q*Ant3,
17, 10, -1, -1, Q*Ant4,
18, 10, -1, -1, Q*Ant5,
]
# Wash1
lwe_model.root.input.model.connections.switch_002.section = 2
lwe_model.root.input.model.connections.switch_002.connections = [0, 1, -1, -1, Q,
1, 2, -1, -1, Q,
2, 4, -1, -1, Q,
3, 4, -1, -1, 0,
4, 5, -1, -1, Q,
5, 6, -1, -1, Q,
6, 7, -1, -1, Q*Ant1,
6, 8, -1, -1, Q*Ant2,
6, 9, -1, -1, Q*Ant3,
6, 11, -1, -1, Q*Ant4,
6, 12, -1, -1, Q*Ant5,
7, 13, -1, -1, Q*Ant1,
8, 13, -1, -1, Q*Ant2,
9, 13, -1, -1, Q*Ant3,
11, 13, -1, -1, Q*Ant4,
12, 13, -1, -1, Q*Ant5,
13, 14, -1, -1, Q*Ant1,
13, 15, -1, -1, Q*Ant2,
13, 16, -1, -1, Q*Ant3,
13, 17, -1, -1, Q*Ant4,
13, 18, -1, -1, Q*Ant5,
14, 10, -1, -1, Q*Ant1,
15, 10, -1, -1, Q*Ant2,
16, 10, -1, -1, Q*Ant3,
17, 10, -1, -1, Q*Ant4,
18, 10, -1, -1, Q*Ant5,
]
# Wash2
lwe_model.root.input.model.connections.switch_003.section = 3
lwe_model.root.input.model.connections.switch_003.connections = [0, 1, -1, -1, Q,
1, 2, -1, -1, Q,
2, 4, -1, -1, Q,
3, 4, -1, -1, 0,
4, 5, -1, -1, Q,
5, 6, -1, -1, Q,
6, 7, -1, -1, Q*Ant1,
6, 8, -1, -1, Q*Ant2,
6, 9, -1, -1, Q*Ant3,
6, 11, -1, -1, Q*Ant4,
6, 12, -1, -1, Q*Ant5,
7, 13, -1, -1, Q*Ant1,
8, 13, -1, -1, Q*Ant2,
9, 13, -1, -1, Q*Ant3,
11, 13, -1, -1, Q*Ant4,
12, 13, -1, -1, Q*Ant5,
13, 14, -1, -1, Q*Ant1,
13, 15, -1, -1, Q*Ant2,
13, 16, -1, -1, Q*Ant3,
13, 17, -1, -1, Q*Ant4,
13, 18, -1, -1, Q*Ant5,
14, 10, -1, -1, Q*Ant1,
15, 10, -1, -1, Q*Ant2,
16, 10, -1, -1, Q*Ant3,
17, 10, -1, -1, Q*Ant4,
18, 10, -1, -1, Q*Ant5,
]
# Elutio
lwe_model.root.input.model.connections.switch_004.section = 4
lwe_model.root.input.model.connections.switch_004.connections = [0, 1, -1, -1, Q,
1, 2, -1, -1, Q,
2, 4, -1, -1, Q,
3, 4, -1, -1, 0,
4, 5, -1, -1, Q,
5, 6, -1, -1, Q,
6, 7, -1, -1, Q*Ant1,
6, 8, -1, -1, Q*Ant2,
6, 9, -1, -1, Q*Ant3,
6, 11, -1, -1, Q*Ant4,
6, 12, -1, -1, Q*Ant5,
7, 13, -1, -1, Q*Ant1,
8, 13, -1, -1, Q*Ant2,
9, 13, -1, -1, Q*Ant3,
11, 13, -1, -1, Q*Ant4,
12, 13, -1, -1, Q*Ant5,
13, 14, -1, -1, Q*Ant1,
13, 15, -1, -1, Q*Ant2,
13, 16, -1, -1, Q*Ant3,
13, 17, -1, -1, Q*Ant4,
13, 18, -1, -1, Q*Ant5,
14, 10, -1, -1, Q*Ant1,
15, 10, -1, -1, Q*Ant2,
16, 10, -1, -1, Q*Ant3,
17, 10, -1, -1, Q*Ant4,
18, 10, -1, -1, Q*Ant5,
]
# Strip
lwe_model.root.input.model.connections.switch_005.section = 5
lwe_model.root.input.model.connections.switch_005.connections = [0, 1, -1, -1, Q,
1, 2, -1, -1, Q,
2, 4, -1, -1, Q,
3, 4, -1, -1, 0,
4, 5, -1, -1, Q,
5, 6, -1, -1, Q,
6, 7, -1, -1, Q*Ant1,
6, 8, -1, -1, Q*Ant2,
6, 9, -1, -1, Q*Ant3,
6, 11, -1, -1, Q*Ant4,
6, 12, -1, -1, Q*Ant5,
7, 13, -1, -1, Q*Ant1,
8, 13, -1, -1, Q*Ant2,
9, 13, -1, -1, Q*Ant3,
11, 13, -1, -1, Q*Ant4,
12, 13, -1, -1, Q*Ant5,
13, 14, -1, -1, Q*Ant1,
13, 15, -1, -1, Q*Ant2,
13, 16, -1, -1, Q*Ant3,
13, 17, -1, -1, Q*Ant4,
13, 18, -1, -1, Q*Ant5,
14, 10, -1, -1, Q*Ant1,
15, 10, -1, -1, Q*Ant2,
16, 10, -1, -1, Q*Ant3,
17, 10, -1, -1, Q*Ant4,
18, 10, -1, -1, Q*Ant5,
]
else:
lwe_model.root.input.model.connections.nswitches = 1
lwe_model.root.input.model.connections.switch_000.section = 0
lwe_model.root.input.model.connections.switch_000.connections = [0, 1, -1, -1, Q,
1, 2, -1, -1, Q,
2, 4, -1, -1, Q,
3, 4, -1, -1, 0,
4, 5, -1, -1, Q,
5, 6, -1, -1, Q,
6, 7, -1, -1, Q*Ant1,
6, 8, -1, -1, Q*Ant2,
6, 9, -1, -1, Q*Ant3,
6, 11, -1, -1, Q*Ant4,
6, 12, -1, -1, Q*Ant5,
7, 13, -1, -1, Q*Ant1,
8, 13, -1, -1, Q*Ant2,
9, 13, -1, -1, Q*Ant3,
11, 13, -1, -1, Q*Ant4,
12, 13, -1, -1, Q*Ant5,
13, 14, -1, -1, Q*Ant1,
13, 15, -1, -1, Q*Ant2,
13, 16, -1, -1, Q*Ant3,
13, 17, -1, -1, Q*Ant4,
13, 18, -1, -1, Q*Ant5,
14, 10, -1, -1, Q*Ant1,
15, 10, -1, -1, Q*Ant2,
16, 10, -1, -1, Q*Ant3,
17, 10, -1, -1, Q*Ant4,
18, 10, -1, -1, Q*Ant5,
]
print(bcolors.OKBLUE + 'no flow changes' + bcolors.ENDC)
#### Execute Simulation ####
## Discretization ##
set_discretization(lwe_model, n_bound=n_comp*[1], n_col=n_column, n_par=n_part)
# Run Simulation
simulation_name_h5 = 'Sim_e084-0007.h5'
run_simulation(lwe_model,simulation_name_h5)
######################
###### Plotting ######
######################
###############################
#### Plot the Buffer Inlet ####
###############################
#fig, ax1 = plt.subplots()
#ax1.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_000.solution_outlet[:,0], 'k', label='Salt')
#ax1.set_xlabel('Time (s)')
#ax1.set_ylabel('Salt Inlet Concentration (mM)')
#ax2 = ax1.twinx()
#ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_000.solution_outlet[:,1], 'r', label='BSA')
#ax2.set_ylabel('BSA Inlet Concentration (mM)')
#lines, labels = ax1.get_legend_handles_labels()
#lines2, labels2 = ax2.get_legend_handles_labels()
#ax2.legend(lines + lines2, labels + labels2)
###############################
#### Plot the Sample Inlet ####
###############################
#fig, ax1 = plt.subplots()
#ax1.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_004.solution_inlet[:,0], 'k', label='Salt')
#ax1.set_xlabel('Time (s)')
#ax1.set_ylabel('Salt Inlet Concentration (mM)')
#ax2 = ax1.twinx()
#ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_004.solution_inlet[:,1], 'r', label='BSA')
#ax2.set_ylabel('BSA Inlet Concentration (mM)')
#lines, labels = ax1.get_legend_handles_labels()
#lines2, labels2 = ax2.get_legend_handles_labels()
#ax1.legend(lines + lines2, labels + labels2)
fig, ax1 = plt.subplots()
fig.patch.set_facecolor('xkcd:white')#('xkcd:mint green')
ax1.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_000.solution_outlet[:,0], 'k', label='Salt')
ax1.set_xlabel('Zeit s')
ax1.set_ylabel('Salt Inlet Concentration (mM)')
ax2 = ax1.twinx()
ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_003.solution_outlet[:,1], 'r', label='pDNA')
ax2.set_ylabel('pDNA Inlet Concentration (mM)')
lines, labels = ax1.get_legend_handles_labels()
plt.savefig('Input Simulation.png', dpi=400)
#salt_step = pandas.read_csv('../01_Input/Natrix_Q_Mini_salt_step.csv')
#input_salt_step = salt_step.to_numpy(dtype=object)
pDNA_exp = pandas.read_csv('../01_Input/Inputdata_P718043-E084-007_NatrixQ_mini.csv')
Salt_exp = pandas.read_csv('../01_Input/InputDataSalt-007.csv')
input_pDNA_exp = pDNA_exp.to_numpy(dtype=object)
input_Salt_exp = Salt_exp.to_numpy(dtype=object)
#############################
#### Plot the simulation ####
#############################
fig, ax1 = plt.subplots()
fig.patch.set_facecolor('xkcd:white')#('xkcd:mint green')
#ax1.plot(lwe_model.root.output.solution.solution_times*Q/(1.66667e-8*60), lwe_model.root.output.solution.unit_008.solution_outlet[:,0], 'k', label='Salt')
ax1.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_010.solution_outlet[:,0], 'k', label='Salt')
#ax1.plot(input_salt_step[:,0],input_salt_step[:,1],'g',label='Salt_Exp')
ax1.set_xlabel('Zeit in s')
ax1.set_ylabel('Salz konz mM')
ax1.set_xlim([2000, 6000])
ax2 = ax1.twinx()
ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_010.solution_outlet[:,1], 'r', label='pDNAMembran2')
ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_013.solution_inlet[:,1], 'y', label='pDNAMembran1')
#ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_007.solution_outlet[:,1]*Ant1, 'r', label='pDNA1')
#ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_008.solution_outlet[:,1]*Ant2, 'y', label='pDNA2')
#ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_009.solution_outlet[:,1]*Ant3, 'm', label='pDNA3')
#ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_011.solution_outlet[:,1]*Ant4, 'c', label='pDNA4')
#ax2.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_012.solution_outlet[:,1]*Ant5, 'peru', label='pDNA5')
ax2.plot(input_pDNA_exp[:,0],input_pDNA_exp[:,1],'b',label='pDNA_Exp')
ax2.set_ylabel('pDNA Outlet Concentration (mM)')
ax3 = ax1.twinx()
ax3.plot(input_Salt_exp[:,0], input_Salt_exp[:,1], 'g', label='Salt_Exp')
#ax4 = ax1.twinx()
#ax4.plot(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_006.soution_solid[:,1], 'g', lable='BeladungMembran2')
lines, labels = ax1.get_legend_handles_labels()
lines2, labels2 = ax2.get_legend_handles_labels()
ax1.legend(lines + lines2, labels + labels2)
plt.savefig('Result_Simulation_P718043-E084-0007.png', dpi=400,bbox_inches='tight')
###################################
###### Saving the Simulation ######
###################################
## Timestamp creation ##
now = datetime.now()
date = datetime.date(now)
hour = now.hour
minute = now.minute
second = now.second
timestamp = str(date)+'-'+str(hour)+'-'+str(minute)+'-'+str(second)+'-'
print(timestamp)
# net simulation time
if second >= second_start:
calculation_time_seconds = second - second_start
else:
calculation_time_seconds = second + 60 - second_start
minute = minute - 1
if minute >= minute_start:
calculation_time_minutes = minute - minute_start
else:
calculation_time_minutes = minute + 60 - minute_start
hour = hour - 1
if hour >= hour_start:
calculation_time_hours = hour - hour_start
else:
calculation_time_hours = hour + 24 - hour_start
calculation_time = str(calculation_time_hours)+':'+str(calculation_time_minutes)+':'+str(calculation_time_seconds)
print('Calculation time: '+ calculation_time)
## Including specifications into the file ##
specs_names = np.array(['timestamp:', 'Operating_Person:', 'used computer:', 'calculation time:', 'cv # m^3:','n_comp # mM:', 'input_reading?:', 'row', 'length_0 # m:', 'length_1 # m:', 'length_2 # m:',
'column_diameter # m:', 'tube_diameter # m:', 'Q_mlmin # m^3/s:', 'col_poros:', 'par_poros:',
'par_radius # m:', 'volume_equi_end # m^3:', 'volume_load_end # m^3:', 'volume_wash1_end # m^3:', 'volume_wash2_end # m^3:', 'volume_elution_end # m^3:', 'volume_strip_end m^3:',
'c_pDNA # mM:', 'c_salt_equi # mM:', 'c_salt_wash1 # mM:', 'c_salt_wash2 # mM:', 'c_salt_elution # mM', 'c_salt_strip # mM :',
'V_CSTR_0 # m^3:', 'V_CSTR_1 # m^3:', 'V_CSTR_2 # m^3:', 'col_disp_0 # m^2/s:', 'col_disp_1 # m^2/s:', 'col_disp_2 # m^2/s:',
'film_diff_2[0] # m/s:', 'film_diff_2[1] # m/s:', 'par_diff_2[0] # m^2/s:',
'par_diff_2[1] # m^2/s:', 'surf_diff_2[0] # m^2/s:', 'surf_diff_2[1] # m^2/s:',
'ka[0] # m^3/s*m^3:', 'ka[1] # m^3/s*m^3:', 'kd[0] # 1/s:', 'kd[1] # 1/s:', 'Nu[0]:', 'Nu[1]:',
'sigma[0]:', 'sigma[1]:', 'lambda_ # mol*m^3:', 'q_init[0] # mM:', 'q_init[1] # mM:', 'n_col', 'n_par'], dtype=object)
specs = np.array([timestamp, Operating_Person, platform.node(), calculation_time, cv, n_comp, str(input_reading), row, length_0, length_1, length_2,
column_diameter, tube_diameter, Q_mlmin, col_poros, par_poros,
par_radius, volume_equi_end, volume_load_end, volume_wash1_end, volume_wash2_end, volume_elution_end, volume_strip_end,
c_pDNA, c_salt_equi, c_salt_load, c_salt_wash1, c_salt_wash2, c_salt_elution, c_salt_strip,
V_CSTR_0, V_CSTR_1, V_CSTR_2, col_disp_0, col_disp_1, col_disp_2, film_diff_2[0], film_diff_2[1],
par_diff_2[0], par_diff_2[1], surf_diff_2[0], surf_diff_2[1],
ka[0], ka[1], kd[0], kd[1], Nu[0], Nu[1],
sigma[0], sigma[1], lambda_, q_init[0], q_init[1], n_column, n_part], dtype=object)
## Elongating the arrays to all be the same size ##
zeros_long = np.zeros((len(lwe_model.root.output.solution.solution_times)))
zeros_0 = np.zeros((1,len(lwe_model.root.output.solution.solution_times)-len(specs)))
specs_names_ext = np.append(specs_names, zeros_0)
specs_ext = np.append(specs, zeros_0)
## For input reading create folders and a file to combine the output data ##
if row == start_row and input_reading:
if saving_csv:
os.mkdir(Path.home()/'CADET_Processes'/'Sheets'/timestamp_start_0)
if combining_output_data:
np.savetxt(Path.home()/'CADET_Processes'/'Sheets'/timestamp_start_0/combined_data_filename,
lwe_model.root.output.solution.solution_times, fmt='%s', delimiter=';', newline='\n',header='time')
if saving_plot:
os.mkdir(Path.home()/'CADET_Processes'/'Plots'/timestamp_start_0)
## Executing save of csv-file ##
if saving_csv:
if input_reading:
save_to_csv_input_reading(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_008.solution_outlet[:,1],
zeros_long, specs_names_ext, specs_ext, row, timestamp_start_0, file_name = timestamp + Simulation_name)
else:
save_to_csv(lwe_model.root.output.solution.solution_times, lwe_model.root.output.solution.unit_008.solution_outlet[:,1],
zeros_long, specs_names_ext, specs_ext, file_name = timestamp + Simulation_name)
else:
print(bcolors.FAIL + 'WARNING: csv-file was not saved' + bcolors.ENDC)
## Executing save of Plot ##
if saving_plot:
if input_reading:
save_plot_input_reading(Simulation_name, timestamp_start_0, timestamp)
else:
save_plot(Simulation_name, timestamp)
else:
print(bcolors.FAIL + 'WARNING: Plot was not saved' + bcolors.ENDC)
## combine the output data into one file ##
if input_reading and saving_csv and combining_output_data:
combine_output(combined_data_filename, Simulation_name,timestamp_start_0, timestamp, row)
## count up, show percentage and total calculation time so far ##
if input_reading:
row = row+1
if stop_row>start_row+1:
percentage = 100*(row-start_row)/(stop_row-start_row)
print(bcolors.OKBLUE + 'Approximately ' + str(round(percentage,2)) +' % done' + bcolors.ENDC)
now = datetime.now()
date = datetime.date(now)
hour = now.hour
minute = now.minute
second = now.second
if second >= second_start_0:
total_calculation_time_seconds_so_far = second - second_start_0
else:
total_calculation_time_seconds_so_far = second + 60 - second_start_0
minute = minute - 1
if minute >= minute_start_0:
total_calculation_time_minutes_so_far = minute - minute_start_0
else:
total_calculation_time_minutes_so_far = minute + 60 - minute_start_0
hour = hour - 1
if hour >= hour_start_0:
total_calculation_time_hours_so_far = hour - hour_start_0
else:
total_calculation_time_hours_so_far = hour + 24 - hour_start_0
total_calculation_time_so_far = str(total_calculation_time_hours_so_far)+':'+str(total_calculation_time_minutes_so_far)+':'+str(total_calculation_time_seconds_so_far)
print(bcolors.OKBLUE+'total calculation time so far: '+ total_calculation_time_so_far + bcolors.ENDC)
print(bcolors.OKGREEN + 'Done' + bcolors.ENDC)