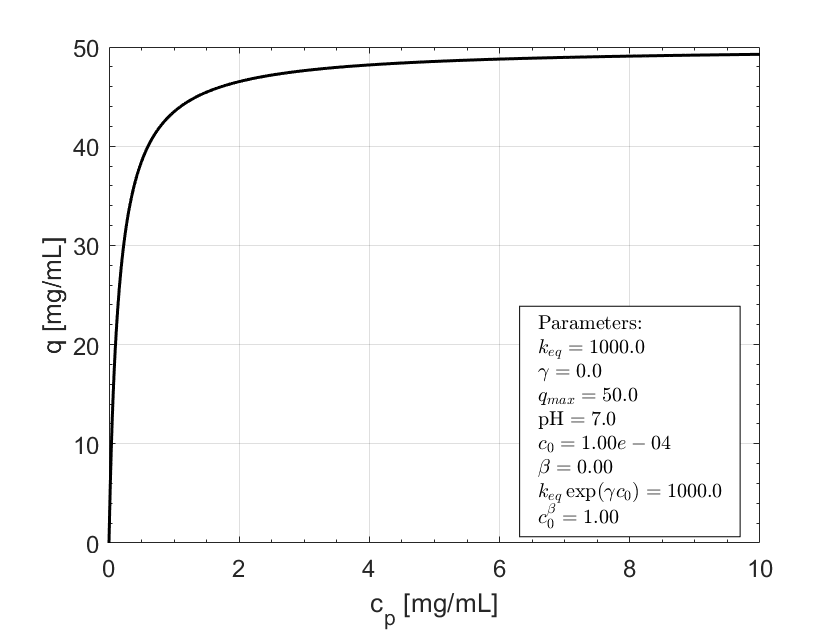
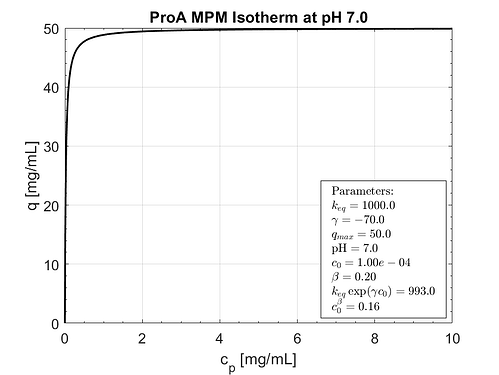
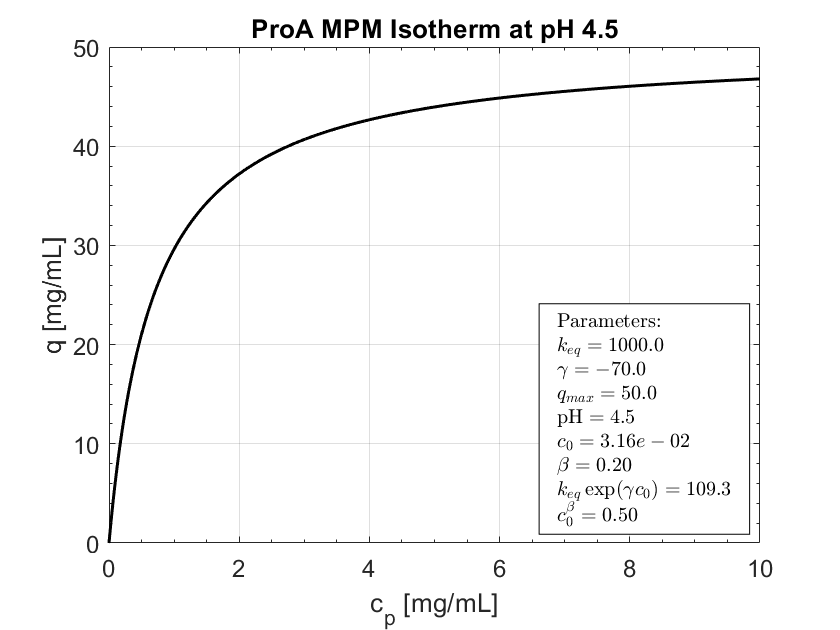
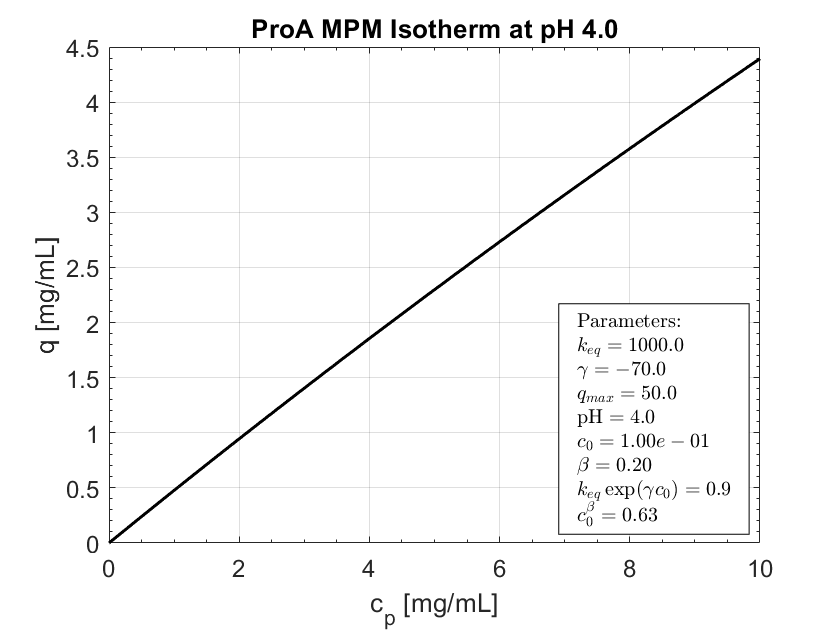
Expected behavior.
Fit Langmuir MPM parameters (offline) to match desired pH dependence. Use fit params to simulate bind, wash, and elute.
Actual behavior
With certain combinations of MPM params (Ka, gamma, beta) , no binding occurs and during elution the outlet concentration dives negative.
How to produce bug (including a minimal reproducible example)
import pandas as pd
import numpy as np
#from numpy import trapezoid
import matplotlib.pyplot as plt
# import cadet packages
from CADETProcess.processModel import ComponentSystem
from CADETProcess.processModel import LangmuirLDF
from CADETProcess.processModel import MobilePhaseModulator
from CADETProcess.processModel import Inlet, LumpedRateModelWithoutPores, Outlet
from CADETProcess.processModel import TubularReactor, Cstr
from CADETProcess.processModel import FlowSheet
from CADETProcess.processModel import Process
from CADETProcess.reference import ReferenceIO
from CADETProcess.comparison import Comparator
from CADETProcess.optimization import OptimizationProblem
from CADETProcess.optimization import U_NSGA3
# --------------------------
# setup experiment
# --------------------------
mLmin_to_m3s = 60*1e6 # conversion
exp_flow = 8. # MV/min (mL/min)
Q = exp_flow/(mLmin_to_m3s) # convert to m^3/s
size_kDa = 150 # protein size
Qmax = 80 # mg/mL = g/L
Qmax_mMol = Qmax/size_kDa
# -------------------------------------
# load and wash and elute volumes and pH
# -------------------------------------
loadVol = 60.1 #42.3 #43.0 #60.1 # mL
washVol = 35.35 #33.9 #25.0 #34.8 # mL
eluteVol = 12. # mL
protein_c = 3.5 #3.7 #3.6 #3.5 # g/L
pH_bind = 7.5
pH_elute = 3.8
pH_b_H = 10**(-pH_bind)
pH_e_H = 10**(-pH_elute)
#print(pH_range)
#print(pH_b_H, 'bind')
#print(pH_e_H,'elute')
# --------------------------------------
# calculating timepoints for experiment
# --------------------------------------
total_vol = loadVol + washVol + eluteVol # mL
exp_time = total_vol/exp_flow*60 # seconds
sample_vol = loadVol
load_time = loadVol/(exp_flow/60.)
wash_time = washVol/(exp_flow/60.)
elute_time = eluteVol/(exp_flow/60.)
# ---------------------------------------------------
# CSTR and tubing lenght parameters from system fit
# ---------------------------------------------------
sys_CSTR_vol = 0.116e-6 # m^3
sys_tubing_len = 3.086 # m
# --------------------------------
# MPM fitting coefficients, keq = Ka * \frac{e^{\gamma*[H+]}}{[H+]^\beta}
# --------------------------------
# !!!!!! varying combinations of these parameters introduces numerical artifacts,
# no bidning, negative elution, etc.... how do I avoid this?
Ka = 1.0e-9 # similar issue with beta
gamma = 0
beta = 2.3 # 2.29 works, not 2.3
# ---------------------------
# create component system
# ---------------------------
component_system = ComponentSystem(['pH','protein'])
# -------------------------
# binding model
# -------------------------
binding_model = MobilePhaseModulator(component_system, name='MPM')
# setup system to use keq = a * np.exp[H+]/[H+]^\beta
binding_model.is_kinetic = False
binding_model.desorption_rate = [0.,1.]
binding_model.capacity = [0,Qmax_mMol]
binding_model.ion_exchange_characteristic = [0, beta]
binding_model.hydrophobicity = [0, gamma]
binding_model.adsorption_rate =[0,Ka]
binding_model.desorption_rate = [0,1]
binding_model.bound_states=[0,1]
binding_model.check_required_parameters()
# -------------------------
# setup BWE buffers
# -------------------------
# feed (mAb05)
feed = Inlet(component_system, name='feed')
feed.c = [pH_b_H,protein_c/size_kDa]
# wash (washout buffer)
wash = Inlet(component_system, name='wash')
wash.c = [pH_b_H,0]
# elute (elution buffer)
elute = Inlet(component_system, name='elute')
elute.c = [pH_e_H,0]
# ----------------------
# device
# ----------------------
device = LumpedRateModelWithoutPores(component_system, name='device')
device.length = 0.0193*8/100 # m
device.diameter = 2.85/100 # m
device.total_porosity = 0.55 # estimate
device.axial_dispersion = 1e-12 # near zero
device.binding_model = binding_model
device.discretization.ncol = 20 # i've gone up to 1000, still an issu
device.c = [pH_b_H,0]
#device.discretization.use_analytic_jacobian = False
# ----------------------
# feed tubing
# ----------------------
load_tubing = TubularReactor(component_system, name='load_tubing')
load_tubing.c = [pH_b_H,0]
load_tubing.length = sys_tubing_len # m #270/100
load_tubing.diameter = 0.75/1000
load_tubing.axial_dispersion = 1e-12
load_tubing.discretization.ncol=20
# ----------------------
# outlet
# ----------------------
outlet = Outlet(component_system, name='outlet')
outlet.solution_recorder.write_solution_bulk = True
# -----------------------
# inlet CSTR
# -----------------------
cstr_vol = 0.18e-6 # m^3
inlet_mixer = Cstr(component_system, name='inlet_mixer')
inlet_mixer.c = [pH_b_H,0]
inlet_mixer.init_liquid_volume = 1.0*cstr_vol # m^3
inlet_mixer.check_required_parameters()
#inlet_mixer.solution_recorder.write_solution_bulk=True
# ------------------------
# outlet CSTR
# ------------------------
outlet_mixer = Cstr(component_system, name='outlet_mixer')
outlet_mixer.c = [pH_b_H,0]
outlet_mixer.init_liquid_volume = 7.0*cstr_vol # m^3
outlet_mixer.check_required_parameters()
#outlet_mixer.solution_recorder.write_solution_bulk=True
# ------------------------
# detector CSTR
# ------------------------
detector_mixer = Cstr(component_system, name='detector_mixer')
detector_mixer.c = [pH_b_H,0]
detector_mixer.init_liquid_volume = 0.2e-6 # m^3 (0.2 mL detector volume)
detector_mixer.check_required_parameters()
# ------------------------
# system CSTR
# ------------------------
system_mixer = Cstr(component_system, name='system_mixer')
system_mixer.c = [pH_b_H,0]
system_mixer.init_liquid_volume = sys_CSTR_vol # m^3
system_mixer.check_required_parameters()
# ------------------------------------------------
# now we create the Flow Sheet
# ------------------------------------------------
flow_sheet = FlowSheet(component_system)
flow_sheet.add_unit(feed)
flow_sheet.add_unit(wash)
flow_sheet.add_unit(elute)
flow_sheet.add_unit(load_tubing)
flow_sheet.add_unit(device)
flow_sheet.add_unit(outlet)
flow_sheet.add_unit(inlet_mixer)
flow_sheet.add_unit(outlet_mixer)
flow_sheet.add_unit(detector_mixer)
flow_sheet.add_unit(system_mixer)
# connect components
flow_sheet.add_connection(feed, system_mixer)
flow_sheet.add_connection(wash, system_mixer)
flow_sheet.add_connection(elute, system_mixer)
flow_sheet.add_connection(system_mixer, load_tubing)
flow_sheet.add_connection(load_tubing, inlet_mixer)
flow_sheet.add_connection(inlet_mixer,device)
flow_sheet.add_connection(device, outlet_mixer)
flow_sheet.add_connection(outlet_mixer, detector_mixer)
flow_sheet.add_connection(detector_mixer, outlet)
# --------------------------------------------------------------
# now we create the process, i.e., turning feed on, swap to wash buffer, same flow rate
# --------------------------------------------------------------
process = Process(flow_sheet, 'breakthrough')
process.add_event('feed_on','flow_sheet.feed.flow_rate',Q,time=0)
process.add_event('feed_off','flow_sheet.feed.flow_rate',0,time=load_time)
# wash
process.add_event('wash_on','flow_sheet.wash.flow_rate',Q,time=load_time)
process.add_event('wash_off','flow_sheet.wash.flow_rate',0,time=load_time+wash_time)
# elute
process.add_event('elute_on','flow_sheet.elute.flow_rate',Q,time=load_time + wash_time)
process.add_event('elute_off','flow_sheet.elute.flow_rate',0,time=exp_time)
# cycle time
process.cycle_time = exp_time # seconds
#process.n_cycles = 1
# -----------------------------
# simluate the process
# -----------------------------
if __name__ == '__main__':
from CADETProcess.simulator import Cadet
process_simulator = Cadet()
process_simulator.time_resolution = 0.1
simulation_results = process_simulator.simulate(process)
# ------------------------------
# save x- and y- axis information
# ------------------------------
x_axis_sim_init = simulation_results.solution.outlet.outlet.time
x_axis_sim_min = x_axis_sim_init/60.
y_axis_sim_init = simulation_results.solution.outlet.outlet.solution
y_axis_sim_init = y_axis_sim_init[:,1]
# ----------------------------
# plot simulation
# ----------------------------
plt.plot(x_axis_sim_min*exp_flow,y_axis_sim_init*size_kDa,'-b',label='BWE simulation')
plt.ylabel('g/L')
plt.xlabel('mL')
plt.legend()
File produced by conda env export > environment.yml
environment.yml (7.56 KB)