Hi all,
I had some general questions involving how CADET employs hyperthreading when performing simulations.
When specifying number of threads in the CADET simulation framework (sim.nThreads), how are the multiple threads used in a single CADET simulation? I was under the impression that the solution in its time and spatial elements was sequential and not parallelizable, however I could be totally off-base here.
Which scenario (out of the below two) would we expect to be more efficient for parameter fitting (assuming the algorithm itself is parallelizable e.g. genetic algorithm)?
- Running 10 simulations in parallel with each simulation containing 1 thread
- Running only 1 simulation at a time with each simulation containing 10 threads
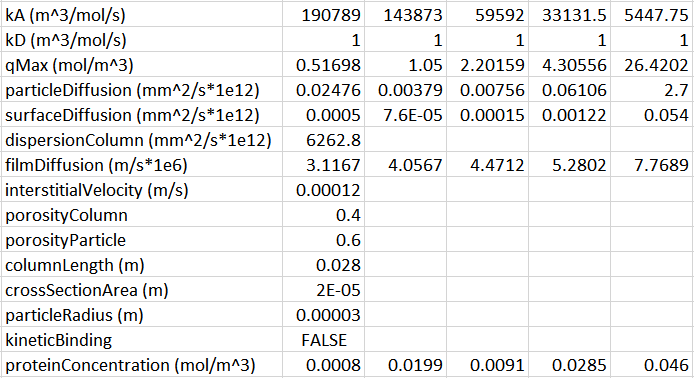
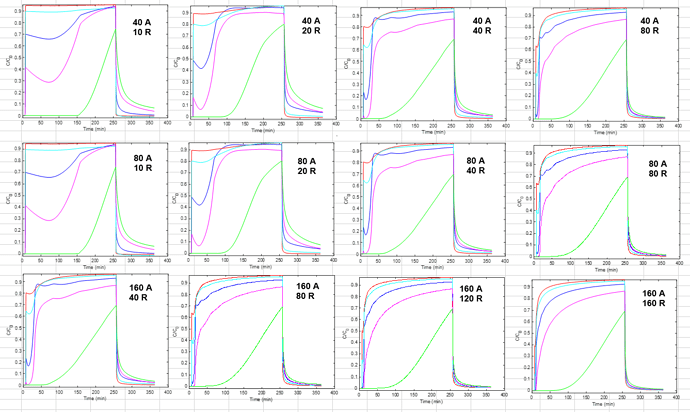
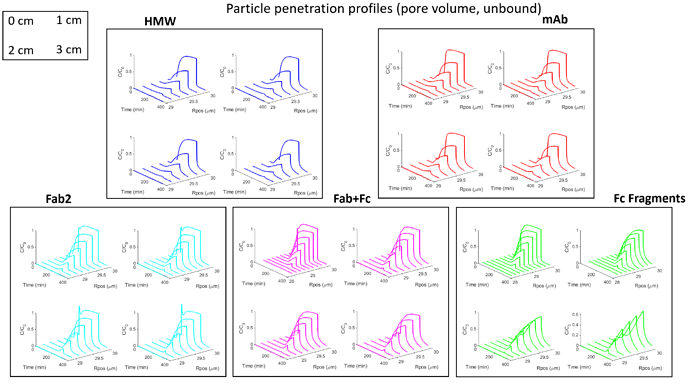
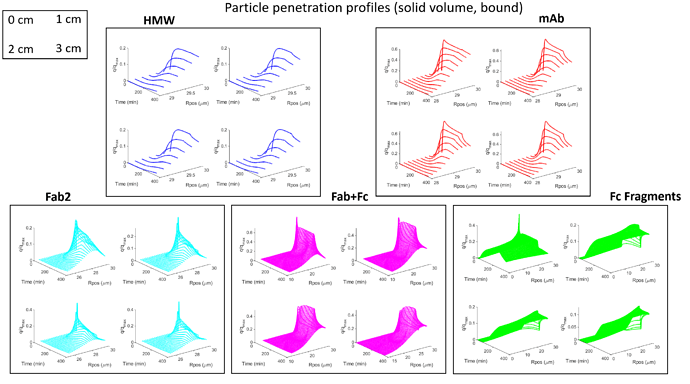
For some context: I am fitting a set of BT curves (single salt/pH, multiple residence times) in CADET MATLAB using the GRM and multicomponent Langmuir isotherm with five protein components. There are many parameters to be fitted here and the simulations are quite slow since I am using a large number of discretizations (150 axial, 100 radial) because there are anomalous fronts in the BT curves that appear otherwise. I have been experimenting with different optimization methods and found that MATLAB surrogateopt (surrogate optimization) is a good option for this problem. I have also spent a considerable amount of time to narrow down the search space of the parameters (kA, qMax, particleDiffusion). The resin in question here has extremely small pores and very strong binding.
Thanks for the help!
Scott