Many model parameters in CADET-Process are not scalar.
For instance, their size depends on the number of components, binding sites, reactions, etc.
CADET-Process provides some functionality to manage these sized parameters, especially while adding events and optimization variables.
Note:
This is still work in progress.
Some things might still not work 100 % as expected.
Some aspects of the interface might change.
If you want to follow the development, check out the PR.
For this tutorial, consider the following process model:
import copy
import numpy as np
from CADETProcess.processModel import ComponentSystem
component_system = ComponentSystem(2)
from CADETProcess.processModel import Inlet
inlet = Inlet(component_system, 'inlet')
from CADETProcess.processModel import Inlet, LumpedRateModelWithPores, Outlet
inlet = Inlet(component_system, name='inlet')
column = LumpedRateModelWithPores(component_system, 'column')
outlet = Outlet(component_system, 'outlet')
from CADETProcess.processModel import FlowSheet, Process
flow_sheet = FlowSheet(component_system)
flow_sheet.add_unit(inlet)
flow_sheet.add_unit(column)
flow_sheet.add_unit(outlet)
flow_sheet.add_connection(inlet, column)
flow_sheet.add_connection(column, outlet)
def setup_process():
process = Process(flow_sheet, 'Demo Indices')
process.cycle_time = 10
return process
from CADETProcess.optimization import OptimizationProblem
def setup_optimization_problem():
optimization_problem = OptimizationProblem('Demo Indices', use_diskcache=False)
optimization_problem.add_evaluation_object(process)
return optimization_problem
Part 1: Convenience functions for setting polynomial parameters
Some parameters in CADET-Process are represented by polynomial coefficients.
For example, the flow rate of an Inlet can be described by a cubic polynomial (in time).
By default, all coefficients are 0.
print(inlet.flow_rate)
[0. 0. 0. 0.]
When assigning a scalar value to the parameter, the state is assumed to be constant.
All other coefficients are set to 0.
Note that polynomial coefficients are specified in order of increasing degree.
I.e. the first coefficient corresponds to the constant term, the second to the linear term, etc.
inlet.flow_rate = 1
print(inlet.flow_rate)
[1. 0. 0. 0.]
It is also possible to specify only any subset of polynomial coefficients.
Again, all unspecified values are assumed to be 0.
inlet.flow_rate = [0, 1]
print(inlet.flow_rate)
[0. 1. 0. 0.]
Or, specify all polynomial coefficients:
inlet.flow_rate = [0, 1, 2, 3]
print(inlet.flow_rate)
[0. 1. 2. 3.]
This also works for parameters with multiple entries, e.g. the inlet concentration.
Set all components to same (constant) concentration:
inlet.c = 1
print(inlet.c)
[[1. 0. 0. 0.]
[1. 0. 0. 0.]]
Specify constant term for each component:
inlet.c = [1, 2]
print(inlet.c)
[[1. 0. 0. 0.]
[2. 0. 0. 0.]]
Specify polynomial coefficients for each component:
inlet.c = [[1, 2], 1]
print(inlet.c)
[[1. 2. 0. 0.]
[1. 0. 0. 0.]]
Specify all polynomial coefficients:
inlet.c = [[0, 1, 2, 3], [4, 5, 6, 7]]
print(inlet.c)
[[0. 1. 2. 3.]
[4. 5. 6. 7.]]
Part 2: Events for multidimensional parameters
When adding events, sometimes, we might not want to change all entries of a parameters but only specific indices.
Just as a reminder, The add_event() method requires the following arguments:
name: Name of the event.parameter_path: Path of the parameter that is changed in dot notation. E.g. to specify theflow_directionparameter of the LumpedRateModelWithPores unit namedcolumn, of the FlowSheet would read asflow_sheet.column.flow_direction.state: Value of the attribute that is changed at Event execution.time: Time at which the event is executed in s.
For example, to add an event called reverse_flow which should modify flow_direction of unit column to value -1 at time 1, and an event called forward_flow which sets the flow_direction back to 1 at time 2, specify the following:
process = setup_process()
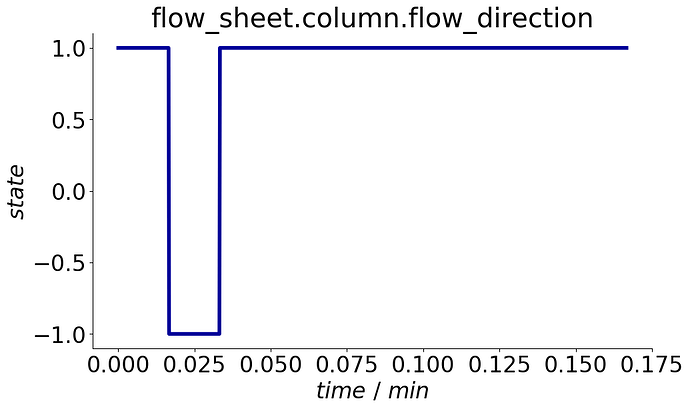
process.add_event('reverse_flow', 'flow_sheet.column.flow_direction', -1, time=1)
print(column.flow_direction)
process.add_event('forward_flow', 'flow_sheet.column.flow_direction', 1, time=2)
print(column.flow_direction)
-1
1
For all non-scalar parameters check the sized_parameters attribute of the Process which aggregates all sized parameters from binding models, units, flow sheet etc.
process.sized_parameters
{'flow_sheet': {'inlet': {'c': array([[0., 0., 0., 0.],
[0., 0., 0., 0.]]),
'flow_rate': array([0., 0., 0., 0.])},
'output_states': {'inlet': [1], 'column': [1], 'outlet': []},
'column': {'film_diffusion': None, 'pore_accessibility': [1, 1]},
'outlet': {}}}
Part 2a: Specify events for 1D arrays
Now, let us consider a 1D parameter. To add an event that modifies all entries of the parameter, you can add an array as state.
E.g., to change the value of the film_diffusion coefficients of the column unit to the value 1e-6 at time=0, add the following:
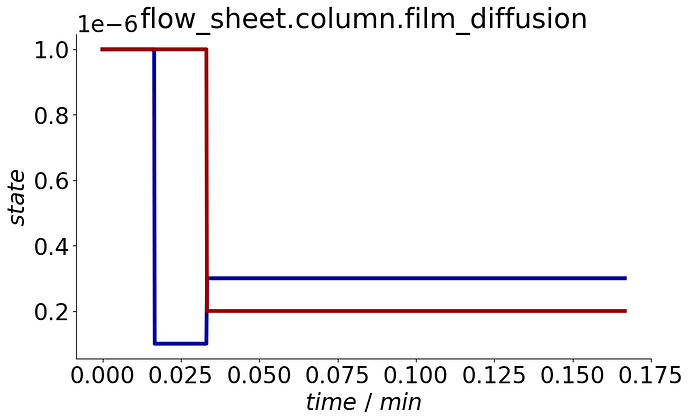
process.add_event(
'film_diffusion_all', 'flow_sheet.column.film_diffusion', [1e-6, 1e-6], time=0
)
print(column.film_diffusion)
[1e-06, 1e-06]
Now, to add an event for single entry in a 1D list / array, add the indices argument.
E.g., to change the value of the film_diffusion coefficient of the first component to 1e-7 at time=1, add indices=0.
process.add_event(
'film_diffusion_index_0', 'flow_sheet.column.film_diffusion', 1e-7, indices=0, time=1
)
print(column.film_diffusion)
[1e-07, 1e-06]
It is also possible to specify multiple indices at once.
The length of indices must match the length of the provided states.
Also, the order of the indices is mapped to the order of the state entries.
process.add_event(
'film_diffusion_index_10', 'flow_sheet.column.film_diffusion', [2e-7, 3e-7], indices=[1, 0], time=2
)
print(column.film_diffusion)
[3e-07, 2e-07]
To visualize the events, a plot_events() method is provided that plots the event state’s evolution over the process cycle.
process.plot_events()
Part 2b: Specify events for 1D polynomial parameters
As mentioned above, polynomial parameters are a bit special with regard to their setter methods.
As with regular 1D parameters, it is possible to specify all state values at once.
To demonstrate this, consider the flow_rate attribute of the inlet unit operation.
process = setup_process()
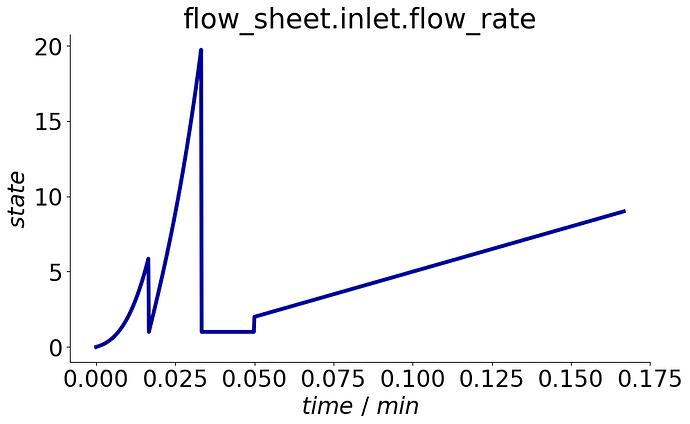
process.add_event(
'flow_rate_all', 'flow_sheet.inlet.flow_rate', [0, 1, 2, 3], time=0
)
print(inlet.flow_rate)
[0. 1. 2. 3.]
Also events that only modify a single entry are equivalent:
process.add_event(
'flow_rate_index_0', 'flow_sheet.inlet.flow_rate', 1, indices=0, time=1
)
print(inlet.flow_rate)
[1. 1. 2. 3.]
However, if no indices are passed, the event acts as if the state is set directly to the parameter.
E.g. setting the state to 1 would automatically fill missing polynomial coefficients and set them to 0.
process.add_event(
'flow_rate_fill_all', 'flow_sheet.inlet.flow_rate', 1, time=2
)
print(inlet.flow_rate)
[1. 0. 0. 0.]
Adding a subset of the coefficients as list also works as expected:
process.add_event(
'flow_rate_fill_subset', 'flow_sheet.inlet.flow_rate', [2, 1], time=3
)
print(inlet.flow_rate)
[2. 1. 0. 0.]
process.plot_events()
Part 2c: Specify events for multidimensional (polynomial) parameters
The process for adding events for multidimensional parameters is similar.
For this purpose, consider the concentration c of the inlet unit operation (which also happens to be polynomial).
process = setup_process()
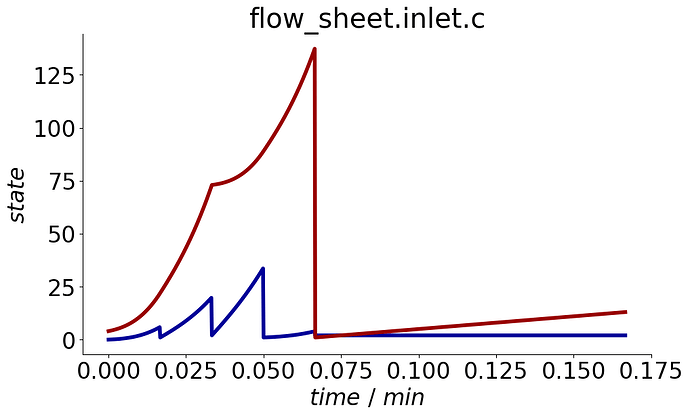
evt = process.add_event(
'c_all', 'flow_sheet.inlet.c',
[[0, 1, 2, 3], [4, 5, 6, 7]],
time=0
)
print(inlet.c)
[[0. 1. 2. 3.]
[4. 5. 6. 7.]]
To add an event that only modify a single entry, make sure that the indices must now be a tuple containig the index of every dimension of the array.
evt = process.add_event(
'c_single', 'flow_sheet.inlet.c', 1, indices=(0, 0), time=1
)
print(inlet.c)
[[1. 1. 2. 3.]
[4. 5. 6. 7.]]
To add multiple indices, make sure to pass a list of tuples.
evt = process.add_event(
'c_multi', 'flow_sheet.inlet.c', [2, 3], indices=[(0, 0), (1, 1)], time=2
)
print(inlet.c)
[[2. 1. 2. 3.]
[4. 3. 6. 7.]]
Slicing is also possible using np.s_.
For more information, refer to this post.
E.g. to add an event that modifies all entries of the first row:
evt = process.add_event(
'c_row', 'flow_sheet.inlet.c', 1, indices=np.s_[0, :], time=3
)
print(inlet.c)
[[1. 1. 1. 1.]
[4. 3. 6. 7.]]
As with 1D polynomials, leveraging the setter convenience function is also possible for polynomial parameters.
E.g. to set the first entry to a constant 2 and the second to value where the constant coefficient is 1 and the linear coefficient is 2 specify the following as state: [2, [1, 2]].
Note, this only works when no indices are provided.
evt = process.add_event(
'c_poly', 'flow_sheet.inlet.c', [2, [1, 2]], time=4
)
print(inlet.c)
[[2. 0. 0. 0.]
[1. 2. 0. 0.]]
process.plot_events()
Part 3: OptimizationVariables
Similar to events, sometimes we only want to add individual entries of a parameter as an optimization variable.
In fact, optimization variables can only be scalar.
Consequently, an index must be provided if the parameter is not scalar.
Just as a reminder, The add_variable() method requires the following arguments:
name: Name of the variable.evaluation_objects: Evaluation object that manages parameter. By default, all evaluation objects are used.parameter_path: Path to access the parameter in the evaluation object.index: Index for variables that modify an entry of a parameter array.
For example, to add a variable bed_porosity that should modify the bed_porosity of unit column, add the following.
Note that the process is automatically added to the optimization problem in the setup method above.
optimization_problem = setup_optimization_problem()
var = optimization_problem.add_variable(
'bed_porosity', evaluation_objects=process, parameter_path='flow_sheet.column.bed_porosity'
)
var.value = 0.5
print(column.bed_porosity)
0.5
Part 3a: Optimizing (multidimensional) arrays
The procedure of adding indices is similar to that of the Event indices.
By default, if no indices are specified, all entries of that array are set to the same value.
E.g. to let the film_diffusion coefficients all have the same value, specify the following:
var = optimization_problem.add_variable(
'film_diffusion_all', evaluation_objects=process, parameter_path='flow_sheet.column.film_diffusion'
)
var.value = 1
print(process.flow_sheet.column.film_diffusion)
[1, 1]
To add a variable that only modifies a single entry of the parameter array, add an indices flag.
E.g. for the first component of the (1D) film_diffusion array:
optimization_problem = setup_optimization_problem()
var = optimization_problem.add_variable(
'film_diffusion_0', evaluation_objects=process, parameter_path='flow_sheet.column.film_diffusion', indices=0
)
var.value = 2
print(process.flow_sheet.column.film_diffusion)
[2.0, 1.0]
Note, for polynomial parameters, if no index is specified, only the constant coefficient is set and the rest of the coefficients are set to zero.
So, to add the constant term for the inlet.flow_rate, use:
var = optimization_problem.add_variable(
'flow_rate_fill', evaluation_objects=process, parameter_path='flow_sheet.inlet.flow_rate'
)
var.value = 1
print(process.flow_sheet.inlet.flow_rate)
[1. 0. 0. 0.]
However, adding indices still works as expected.
E.g. for the linear coefficient of the flow_rate:
optimization_problem = setup_optimization_problem()
var = optimization_problem.add_variable(
'flow_rate_single', evaluation_objects=process, parameter_path='flow_sheet.inlet.flow_rate', indices=1
)
var.value = 2
print(process.flow_sheet.inlet.flow_rate)
[1. 2. 0. 0.]
Similarly, for regular multidimensional arrays, if no indices are specified, the value is set to all entries of that array.
Similarly, multidimensional parameters can be indexed by specifying a tuple.
For example, to optimize the linear coefficient of the first component of the inlet concentration:
optimization_problem = setup_optimization_problem()
var = optimization_problem.add_variable(
'concentration_fill_values_all', evaluation_objects=process, parameter_path='flow_sheet.inlet.c'
)
var.value = 1
print(process.flow_sheet.inlet.c)
[[1. 0. 0. 0.]
[1. 0. 0. 0.]]
Modify const. coefficient of single entry.
optimization_problem = setup_optimization_problem()
var = optimization_problem.add_variable(
'concentration_fill_values_single', evaluation_objects=process, parameter_path='flow_sheet.inlet.c', indices=0
)
var.value = 2
print(process.flow_sheet.inlet.c)
[[2. 0. 0. 0.]
[1. 0. 0. 0.]]
Explicitly modify linear coefficient of single entry.
optimization_problem = setup_optimization_problem()
var = optimization_problem.add_variable(
'concentration_single_entry', evaluation_objects=process, parameter_path='flow_sheet.inlet.c', indices=(0, 1)
)
var.value = 3
print(process.flow_sheet.inlet.c)
[[2. 3. 0. 0.]
[1. 0. 0. 0.]]
Part 3b: Optimizing multidimensional Event states
In certain scenarios, optimizing the state of an event becomes essential.
Depending on which parameter dimensions the event modifies, and the indices chosen for the event state, it might be imperative to include indices in the optimization variable as well.
Consider the slope of an elution gradient that starts at a specific time.
Here, the first component starts at c = 0~mM with a slope of 1~mM / s whereas the second component’s concentration is always 0~mM.
process = setup_process()
optimization_problem = setup_optimization_problem()
evt = process.add_event(
'c_poly', 'flow_sheet.inlet.c', [[0, 1], 0], time=1
)
print(inlet.c)
[[0. 1. 0. 0.]
[0. 0. 0. 0.]]
To add an optimization variable that only modifies the linear coefficient, add the event state as parameter_path and the corresponding index.
var = optimization_problem.add_variable(
'concentration_single_entry', evaluation_objects=process, parameter_path='c_poly.state', indices=(0, 1)
)
var.value = 2
print(inlet.c)
[[0. 2. 0. 0.]
[0. 0. 0. 0.]]
As always, testing and feedback is highly appreciated! ![]()